Groundwater 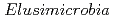 are metabolically diverse compared to gut microbiome
are metabolically diverse compared to gut microbiome 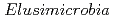 and some have a novel nitrogenase paralog
and some have a novel nitrogenase paralog
M heust, R.*; Castelle, C. J.*; Matheus Carnevali, P. B.*; Farag, I. F.*; He, C.*; Chen, L.-X.*; Amano, Yuki
heust, R.*; Castelle, C. J.*; Matheus Carnevali, P. B.*; Farag, I. F.*; He, C.*; Chen, L.-X.*; Amano, Yuki 

 ; Hug, L. A.*; Banfield, J. F.*
; Hug, L. A.*; Banfield, J. F.*
We reconstructed 94 draft-quality, non-redundant genomes from diverse animal-associated and natural environments. Genomes group into 12 clades, 10 of which previously lacked reference genomes. Groundwater-associated 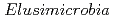 are predicted to be capable of heterotrophic or autotrophic lifestyles, reliant on oxygen or nitrate/nitrite-dependent respiration of fatty acids, or a variety of organic compounds and Rnf-dependent acetogenesis with hydrogen and carbon dioxide as the substrates. Genomes from two clades of groundwater-associated
are predicted to be capable of heterotrophic or autotrophic lifestyles, reliant on oxygen or nitrate/nitrite-dependent respiration of fatty acids, or a variety of organic compounds and Rnf-dependent acetogenesis with hydrogen and carbon dioxide as the substrates. Genomes from two clades of groundwater-associated 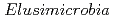 often encode a new homologous group of nitrogenase-like proteins that co-occur with an extensive suite of radical SAM-based proteins. We identified similar genomic loci in genomes of bacteria from the Gracilibacteria and Myxococcus phyla and predict that the gene clusters reduce a tetrapyrrole, possibly to form a novel cofactor. The animal-associated
often encode a new homologous group of nitrogenase-like proteins that co-occur with an extensive suite of radical SAM-based proteins. We identified similar genomic loci in genomes of bacteria from the Gracilibacteria and Myxococcus phyla and predict that the gene clusters reduce a tetrapyrrole, possibly to form a novel cofactor. The animal-associated 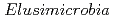 clades nest phylogenetically within two groundwater-associated clades. Thus, we propose an evolutionary trajectory in which some
clades nest phylogenetically within two groundwater-associated clades. Thus, we propose an evolutionary trajectory in which some 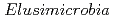 adapted to animal-associated lifestyles from groundwater-associated species via genome reduction.
adapted to animal-associated lifestyles from groundwater-associated species via genome reduction.