Mutagenic potential of clustered DNA damage site in 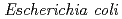
Shikazono, Naoya; Pearson, C.*; Thacker, J.*; O'Neill, P.*
Clustered DNA damage induced by a single radiation track is a unique feature of ionizing radiation. Recent 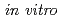 studies have shown that the repair of lesions within clusters may be retarded, but less is known about the processing and the mutagenic effects of such clustered damage in vivo. Using a bacterial plasmid-based assay, we have investigated the mutagenic potential of bistranded clustered damage sites which consist of 8-oxo-7,8-dihydroguanine (8-oxoG) and dihydrothymine (DHT) at defined separations. We found a significantly higher mutation frequency for the clustered DHT + 8-oxoG lesions than that for either a single 8-oxoG or a single DHT in wild-type and in glycosylase-deficient strains of
studies have shown that the repair of lesions within clusters may be retarded, but less is known about the processing and the mutagenic effects of such clustered damage in vivo. Using a bacterial plasmid-based assay, we have investigated the mutagenic potential of bistranded clustered damage sites which consist of 8-oxo-7,8-dihydroguanine (8-oxoG) and dihydrothymine (DHT) at defined separations. We found a significantly higher mutation frequency for the clustered DHT + 8-oxoG lesions than that for either a single 8-oxoG or a single DHT in wild-type and in glycosylase-deficient strains of 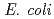 . From these results and similarities with the mutability of respective 8-oxoG + AP clusters, it is suggested that removal of 8-oxoG within clustered damage site is retarded, probably reflecting the preferential excision of DHT initially. For a certain fraction of clusters, 8-oxoG may be initially removed from the cluster. To gain further insights on the processing of the DHT + 8-oxoG cluster, several potential intermediates after 8-oxoG removal were assessed for their mutability. For instance, DHT + AP or DHT + Gap containing cluster, but not AP + AP or Gap +AP clusters, has a relatively low mutation frequency. Further, AP + AP or Gap + AP cluster had a reduced transformation efficiency. These results led us to suggest that, when either 8-oxoG or DHT is initially excised from a cluster containing 8-oxoG and DHT, the base remaining within the resulting damage will not be further converted to an AP site or to a single strand break
. From these results and similarities with the mutability of respective 8-oxoG + AP clusters, it is suggested that removal of 8-oxoG within clustered damage site is retarded, probably reflecting the preferential excision of DHT initially. For a certain fraction of clusters, 8-oxoG may be initially removed from the cluster. To gain further insights on the processing of the DHT + 8-oxoG cluster, several potential intermediates after 8-oxoG removal were assessed for their mutability. For instance, DHT + AP or DHT + Gap containing cluster, but not AP + AP or Gap +AP clusters, has a relatively low mutation frequency. Further, AP + AP or Gap + AP cluster had a reduced transformation efficiency. These results led us to suggest that, when either 8-oxoG or DHT is initially excised from a cluster containing 8-oxoG and DHT, the base remaining within the resulting damage will not be further converted to an AP site or to a single strand break 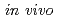 .
.