Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
天野 由記; Sachdeva, R.*; Gittins, D.*; Anantharaman, K.*; Lei, S.*; Valentin-Alvarado, L. E.*; Diamond, S.*; 別部 光里*; 岩月 輝希; 望月 陽人; et al.
Environmental Microbiome (Internet), 19, p.105_1 - 105_17, 2024/12
被引用回数:0 パーセンタイル:0.00(Genetics & Heredity)Underground research laboratories (URLs) provide a window on the deep biosphere and enable investigation of potential microbial impacts on nuclear waste, CO and H
and H stored in the subsurface. We carried out the first multi-year study of groundwater microbiomes sampled from defined intervals between 140 and 400 m below the surface of the Horonobe and Mizunami URLs, Japan. The Horonobe and Mizunami microbiomes are dissimilar, likely because the Mizunami URL is hosted in granitic rock and the Horonobe URL in sedimentary rock. Despite this, hydrogen metabolism, rubisco-based CO
stored in the subsurface. We carried out the first multi-year study of groundwater microbiomes sampled from defined intervals between 140 and 400 m below the surface of the Horonobe and Mizunami URLs, Japan. The Horonobe and Mizunami microbiomes are dissimilar, likely because the Mizunami URL is hosted in granitic rock and the Horonobe URL in sedimentary rock. Despite this, hydrogen metabolism, rubisco-based CO fixation, reduction of nitrogen compounds and sulfate reduction are well represented functions in microbiomes from both URLs, although methane metabolism is more prevalent at the organic- and CO
fixation, reduction of nitrogen compounds and sulfate reduction are well represented functions in microbiomes from both URLs, although methane metabolism is more prevalent at the organic- and CO -rich Horonobe URL. We detected near-identical genotypes for approximately one third of all genomically defined organisms at multiple depths within the Horonobe URL. This cannot be explained by inactivity, as in situ growth was detected for some bacteria, albeit at slow rates. Given the current low hydraulic conductivity and groundwater compositional heterogeneity, ongoing inter-site strain dispersal seems unlikely. Alternatively, the Horonobe URL microbiome homogeneity may be explained by higher groundwater mobility during the last glacial period. Genotypically-defined species closely related to those detected in the URLs were identified in three other subsurface environments in the USA. Thus, dispersal rates between widely separated underground sites may be fast enough relative to mutation rates to have precluded substantial divergence in species composition. Species overlaps between subsurface locations on different continents constrain expectations regarding the scale of global subsurface biodiversity. Overall, microbiome and geochemical stability over the study period has important implications for underground storage applications.
-rich Horonobe URL. We detected near-identical genotypes for approximately one third of all genomically defined organisms at multiple depths within the Horonobe URL. This cannot be explained by inactivity, as in situ growth was detected for some bacteria, albeit at slow rates. Given the current low hydraulic conductivity and groundwater compositional heterogeneity, ongoing inter-site strain dispersal seems unlikely. Alternatively, the Horonobe URL microbiome homogeneity may be explained by higher groundwater mobility during the last glacial period. Genotypically-defined species closely related to those detected in the URLs were identified in three other subsurface environments in the USA. Thus, dispersal rates between widely separated underground sites may be fast enough relative to mutation rates to have precluded substantial divergence in species composition. Species overlaps between subsurface locations on different continents constrain expectations regarding the scale of global subsurface biodiversity. Overall, microbiome and geochemical stability over the study period has important implications for underground storage applications.
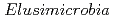 are metabolically diverse compared to gut microbiome
are metabolically diverse compared to gut microbiome 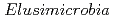 and some have a novel nitrogenase paralog
and some have a novel nitrogenase paralogM heust, R.*; Castelle, C. J.*; Matheus Carnevali, P. B.*; Farag, I. F.*; He, C.*; Chen, L.-X.*; 天野 由記; Hug, L. A.*; Banfield, J. F.*
heust, R.*; Castelle, C. J.*; Matheus Carnevali, P. B.*; Farag, I. F.*; He, C.*; Chen, L.-X.*; 天野 由記; Hug, L. A.*; Banfield, J. F.*
ISME Journal, 14(12), p.2907 - 2922, 2020/12
被引用回数:71 パーセンタイル:96.30(Ecology)We reconstructed 94 draft-quality, non-redundant genomes from diverse animal-associated and natural environments. Genomes group into 12 clades, 10 of which previously lacked reference genomes. Groundwater-associated 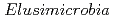 are predicted to be capable of heterotrophic or autotrophic lifestyles, reliant on oxygen or nitrate/nitrite-dependent respiration of fatty acids, or a variety of organic compounds and Rnf-dependent acetogenesis with hydrogen and carbon dioxide as the substrates. Genomes from two clades of groundwater-associated
are predicted to be capable of heterotrophic or autotrophic lifestyles, reliant on oxygen or nitrate/nitrite-dependent respiration of fatty acids, or a variety of organic compounds and Rnf-dependent acetogenesis with hydrogen and carbon dioxide as the substrates. Genomes from two clades of groundwater-associated 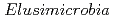 often encode a new homologous group of nitrogenase-like proteins that co-occur with an extensive suite of radical SAM-based proteins. We identified similar genomic loci in genomes of bacteria from the Gracilibacteria and Myxococcus phyla and predict that the gene clusters reduce a tetrapyrrole, possibly to form a novel cofactor. The animal-associated
often encode a new homologous group of nitrogenase-like proteins that co-occur with an extensive suite of radical SAM-based proteins. We identified similar genomic loci in genomes of bacteria from the Gracilibacteria and Myxococcus phyla and predict that the gene clusters reduce a tetrapyrrole, possibly to form a novel cofactor. The animal-associated 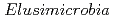 clades nest phylogenetically within two groundwater-associated clades. Thus, we propose an evolutionary trajectory in which some
clades nest phylogenetically within two groundwater-associated clades. Thus, we propose an evolutionary trajectory in which some 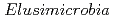 adapted to animal-associated lifestyles from groundwater-associated species via genome reduction.
adapted to animal-associated lifestyles from groundwater-associated species via genome reduction.
Al-Shayeb, B.*; Sachdeva, R.*; Chen, L.-X.*; Ward, F.*; Munk, P.*; Devoto, A.*; Castelle, C. J.*; Olm, M. R.*; Bouma-Gregson, K.*; 天野 由記; et al.
Nature, 578(7795), p.425 - 431, 2020/02
被引用回数:295 パーセンタイル:99.46(Multidisciplinary Sciences)Phage typically have small genomes and depend on their bacterial hosts for replication. We generated metagenomic datasets from many diverse ecosystems and reconstructed hundreds of huge phage genomes, between 200 kbp and 716 kbp in length. Thirty four genomes were manually curated to completion, including the largest phage genomes yet reported. Expanded genetic repertoires include diverse and new CRISPR-Cas systems, tRNAs, tRNA synthetases, tRNA modification enzymes, initiation and elongation factors and ribosomal proteins. Phage CRISPR have the capacity to silence host transcription factors and translational genes, potentially as part of a larger interaction network that intercepts translation to redirect biosynthesis to phage-encoded functions. Some phage repurpose bacterial systems for phage-defense to eliminate competing phage. We phylogenetically define seven major clades of huge phage from human and other animal microbiomes, oceans, lakes, sediments, soils and the built environment. We conclude that large gene inventories reflect a conserved biological strategy, observed across a broad bacterial host range and resulting in the distribution of huge phage across Earth's ecosystems.
Matheus Carnevali, P. B.*; Schulz, F.*; Castelle, C. J.*; Kantor, R. S.*; Shih, P.*; Sharon, I.*; Santini, J.*; Olm, M. R.*; 天野 由記; Thomas, B. C.*; et al.
Nature Communications (Internet), 10, p.463_1 - 463_15, 2019/01
被引用回数:48 パーセンタイル:88.33(Multidisciplinary Sciences)The metabolic platform in which microbial aerobic respiration evolved is tightly linked to the origins of Cyanobacteria (Oxyphotobacteria). Melainabacteria and Sericytochromatia, close phylogenetic neighbores to Oxyphotobacteria comprise both fermentative and aerobic representatives, or clades that are capablee of both. Here, we predict the metabolisms of Margulisbacteria from two distinct environments and Saganbacteria, and compare them to genomes of organisms from the related lineages. Melainabacteria BJ4A obtained from Mizunami site are potentially able to use O and other terminal electron acceptors. The type C heme-copper oxygen reductase found in Melainabacteria BJ4A may be adapted to low O
and other terminal electron acceptors. The type C heme-copper oxygen reductase found in Melainabacteria BJ4A may be adapted to low O levels, as expected for microaerophilic or anoxic environments such as the subsurface. Notably, Melainabacteria BJ4A seems to have a branched electron transport chain, with one branch leading to a cytochrome d ubiquinol oxidoreductase and the other one leading to the type C heme-copper oxygen reductase. Both these enzymes have high affinity for O
levels, as expected for microaerophilic or anoxic environments such as the subsurface. Notably, Melainabacteria BJ4A seems to have a branched electron transport chain, with one branch leading to a cytochrome d ubiquinol oxidoreductase and the other one leading to the type C heme-copper oxygen reductase. Both these enzymes have high affinity for O , thus are adapted to low O
, thus are adapted to low O levels. These contemporary lineages have representatives with fermentative H
levels. These contemporary lineages have representatives with fermentative H -based metabolism, lineages capable of aerobic or anaerobic respiration, and lineages with both. Our findings support the idea that the ancestor of these lineages was an anaerobe in which fermentation and H
-based metabolism, lineages capable of aerobic or anaerobic respiration, and lineages with both. Our findings support the idea that the ancestor of these lineages was an anaerobe in which fermentation and H metabolism were central metabolic features.
metabolism were central metabolic features.
Hug, L. A.*; Baker, B. J.*; Anantharaman, K.*; Brown, C. T.*; Probst, A. J.*; Castelle, C. J.*; Butterfield, C. N.*; Hernsdorf, A. W.*; 天野 由記; 伊勢 孝太郎; et al.
Nature Microbiology (Internet), 1(5), p.16048_1 - 16048_6, 2016/05
被引用回数:1378 パーセンタイル:99.96(Microbiology)生命の系統樹は生物学において最も重要な中心テーマの一つである。遺伝子調査によると、莫大な数のブランチの存在が示唆されているが、フルスケールに近い系統樹でさえわかりにくいのが現状である。本研究では、これまでに報告されてきた配列情報に加えて、新たに取得した未培養生物のゲノム情報を用いて、バクテリア,アーキア,真核生物を含む系統樹を更新した。系統樹の描写は、全体的な概容とそれぞれの主要な系統における多様性のスナップショットの両方について行った。その結果、バクテリアの多様化の優勢性が示され、培養されていない生物種の重要性とともに主要な放射構造においてそれらの生物種の重要な進化が集中している現象が強調された。
 fixation by abundant Nitrospirae in the deep subsurface
fixation by abundant Nitrospirae in the deep subsurface天野 由記; Anantharaman, K.*; Tomas, B. C.*; Olm, M.*; Burstein, D.*; Castelle, C. J.*; 別部 光里*; 宮川 和也; 岩月 輝希; 鈴木 庸平*; et al.
no journal, ,
The bacterial phylum Nitrospirae is phylogenetically diverse. There are relatively few isolated representatives available for laboratory study and the physiology, functions and distributions of these bacteria across environments remain largely unknown. To understand the ecological role of Nitrospirae in the deep subsurface, we analyzed metagenomically-derived near complete genomes from groundwaters associated with granite and sedimentary rock where Nitrospirae are very abundant. The bacteria are autotrophs that fix CO via the Wood-Ljungdahl pathway and reductive TCA cycles. The genomes encode versatile energy-generating pathways that involve sulfate reduction, hydrogen oxidation and nitrite reduction. Phylogenetic analyses indicate that the organisms are most similar to the isolated magnetotactic bacterium, Candidatus Magnetobacterium bavaricum, with only 89-91% 16S rRNA gene sequence identity. These Nitrospirae bacteria appear to play critical ecosystem roles as primary producers and they are likely central to sulfur cycling in the deep subsurface.
via the Wood-Ljungdahl pathway and reductive TCA cycles. The genomes encode versatile energy-generating pathways that involve sulfate reduction, hydrogen oxidation and nitrite reduction. Phylogenetic analyses indicate that the organisms are most similar to the isolated magnetotactic bacterium, Candidatus Magnetobacterium bavaricum, with only 89-91% 16S rRNA gene sequence identity. These Nitrospirae bacteria appear to play critical ecosystem roles as primary producers and they are likely central to sulfur cycling in the deep subsurface.