Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
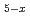 Pt
Pt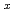
MacLaughlin, D. E.*; Rose, M. S.*; Bernal, O. O.*; Heffner, R. H.; Nieuwenhuys, G. J.*; Chau, R.*; Maple, M. B.*
Physica B; Condensed Matter, 374-375, p.174 - 176, 2006/03
Times Cited Count:0 Percentile:0(Physics, Condensed Matter)Transverse-field 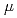 SR shifts and relaxation rates have been measured in the non-Fermi liquid alloy system UCu
SR shifts and relaxation rates have been measured in the non-Fermi liquid alloy system UCu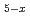 Pt
Pt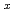 . At low temperatures the fractional spread in Knight shifts is 2 for x=1, but is only half this value for x=1.5 and 2.5. In a disorder-driven scenario where the NFL behavior is due to a broadly distributed characteristic energy E, our results indicate that
. At low temperatures the fractional spread in Knight shifts is 2 for x=1, but is only half this value for x=1.5 and 2.5. In a disorder-driven scenario where the NFL behavior is due to a broadly distributed characteristic energy E, our results indicate that 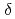 E/E
E/E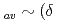 K/K)
K/K)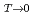 is similar for UCu
is similar for UCu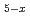 Pd
Pd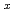 . Our results correlate with the observed suppression of other NFL anomaliess.
. Our results correlate with the observed suppression of other NFL anomaliess.
Okazaki, Nobuo; Ohara, Takashi; Umino, Hisao*; Chatake, Toshiyuki*; Kurihara, Kazuo; Cachau, R. E.*; Blaber, M.*; Niimura, Nobuo*; Kuroki, Ryota
no journal, ,
Okazaki, Nobuo; Ohara, Takashi; Umino, Hisao*; Chatake, Toshiyuki*; Kurihara, Kazuo; Cachau, R. E.*; Blaber, M.*; Niimura, Nobuo*; Kuroki, Ryota
no journal, ,
In protein molecules, key energetic contributors are solvation, desolvation and hydrogen bonding. They contribute protein folding, dynamics and molecular recognition. As a result, more elaborate studies of hydrogen atoms will be great help to recognize protein structures and obtain new findings of them. However, we do not have system which dedicated to characterization and analysis of hydrogen bonding. Therefore, we have developed a database for hydrogen and hydration water molecules. That database named Hydrogen and Hydration Database for Biomolecules (HHDB). Hydrogen bond data stored to HHDB use hydrogen atom coordinates determined directly by neutron diffraction and certain extremely high resolution X-ray diffraction. HHDB provides graphical user interface, users can use it through web browser. HHDB can visualize hydrogen atom positions in protein and solvent, and hydrogen bonding interactions. Fig. 1 shows HHDB plot example. In this plot, hydrogen atom is placed at the origin, and each point represents hydrogen bond distance and angle. We are improving the web user interfaces and the performance for usability.