Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Hiromoto, Takeshi; Adachi, Motoyasu; Shibazaki, Chie; Schrader, T. E.*; Ostermann, A.*; Kuroki, Ryota
JPS Conference Proceedings (Internet), 8, p.033003_1 - 033003_6, 2015/09
T4 phage lysozyme (T4L) is an endoacetylmuramidase that degrades the murein of the bacterial cell wall by cleaving the  -1,4-glycosidic bond between
-1,4-glycosidic bond between 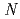 -acetylmuramic acid and
-acetylmuramic acid and 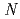 -acetylglucosamine. We previously reported that the substitution of the catalytic Thr26 with the nucleophilic His converts the wild-type (WT) T4L from an inverting to a retaining glycosidase, in which the
-acetylglucosamine. We previously reported that the substitution of the catalytic Thr26 with the nucleophilic His converts the wild-type (WT) T4L from an inverting to a retaining glycosidase, in which the  -configuration of the substrate is retained in the product. We also found that the Thr26His (T26H) mutant can catalyze a transglycosylation reaction more effectively than hydrolysis, although the WT-T4L has no transglycosidase activity. To clarify the role of the substituted His26 in transglycosylation and to investigate its relationship to the neighboring acidic residue Asp20 using neutron crystallography, a perdeuterated recombinant protein of the T26H mutant (d-T26H) was prepared for crystallization. The perdeuterated form was produced in
-configuration of the substrate is retained in the product. We also found that the Thr26His (T26H) mutant can catalyze a transglycosylation reaction more effectively than hydrolysis, although the WT-T4L has no transglycosidase activity. To clarify the role of the substituted His26 in transglycosylation and to investigate its relationship to the neighboring acidic residue Asp20 using neutron crystallography, a perdeuterated recombinant protein of the T26H mutant (d-T26H) was prepared for crystallization. The perdeuterated form was produced in 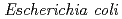 cells cultured in deuterated-rich media. After purification, the d-T26H mutant was crystallized under deuterated conditions and grown to a volume of 0.12 mm
cells cultured in deuterated-rich media. After purification, the d-T26H mutant was crystallized under deuterated conditions and grown to a volume of 0.12 mm using a macroseeding technique. A preliminary neutron-diffraction experiment at 100 K at the FRM II research reactor (Munich, Germany) gave diffraction spots of up to 2.8
using a macroseeding technique. A preliminary neutron-diffraction experiment at 100 K at the FRM II research reactor (Munich, Germany) gave diffraction spots of up to 2.8  resolution after a 1.5 h exposure.
resolution after a 1.5 h exposure.
 -glucosyltransferase from
-glucosyltransferase from 
Hiromoto, Takeshi; Honjo, Eijiro*; Noda, Hisanobu*; Tamada, Taro; Kazuma, Kohei*; Suzuki, Masahiko*; Blaber, M.; Kuroki, Ryota
Protein Science, 24(3), p.395 - 407, 2015/03
Times Cited Count:72 Percentile:89.80(Biochemistry & Molecular Biology)UDP-glucose: anthocyanidin 3- -glucosyltransferase (UGT78K6) from
-glucosyltransferase (UGT78K6) from  catalyzes the transfer of glucose from UDP-glucose to anthocyanidins such as delphinidin. To understand the acceptor-recognition scheme of UGT78K6, the crystal structure of UGT78K6 and its complex forms with anthocyanidin delphinidin and petunidin, and flavonol kaempferol were determined to resolutions of 1.85
catalyzes the transfer of glucose from UDP-glucose to anthocyanidins such as delphinidin. To understand the acceptor-recognition scheme of UGT78K6, the crystal structure of UGT78K6 and its complex forms with anthocyanidin delphinidin and petunidin, and flavonol kaempferol were determined to resolutions of 1.85  , 2.55
, 2.55  , 2.70
, 2.70  and 1.75
and 1.75  respectively. The anthocyanidin- and flavonol-acceptor binding details are almost identical in each complex structure, although the glucosylation activities against each acceptor were significantly different. The acceptor substrates in UGT78K6 are reversely bound to its binding site by a 180
respectively. The anthocyanidin- and flavonol-acceptor binding details are almost identical in each complex structure, although the glucosylation activities against each acceptor were significantly different. The acceptor substrates in UGT78K6 are reversely bound to its binding site by a 180 rotation about the O1-O3 axis of the flavonoid backbones observed in
rotation about the O1-O3 axis of the flavonoid backbones observed in 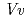 GT1 and UGT78G1. These substrate recognition schemes suggest the potential for controlled synthesis of natural pigments.
GT1 and UGT78G1. These substrate recognition schemes suggest the potential for controlled synthesis of natural pigments.
Adachi, Motoyasu; Arai, Shigeki; Hiromoto, Takeshi; Kuroki, Ryota
Hamon, 24(1), p.45 - 49, 2014/02
Protein structure analysis using neutron diffraction (neutron protein crystallography; NPC) is gaining greater importance in the understanding of structure and function relationships of biological macromolecules such as proteins and DNA. Current developments of neutron diffractometers installed at the JAEA research reactor and pulsed neutron source permit observation of the locations of hydrogen atoms and hydrating water molecules and help understanding of important mechanisms of chemical reactions catalyzed by biological macromolecules. Here, we introduce practical approaches of NPC including sample preparation, crystal growth, structure determination and utilization of information obtained from NPC.
 -glucosyltransferase from
-glucosyltransferase from 
Hiromoto, Takeshi; Honjo, Eijiro*; Tamada, Taro; Noda, Hisanobu*; Kazuma, Kohei*; Suzuki, Masahiko*; Kuroki, Ryota
Journal of Synchrotron Radiation, 20(6), p.894 - 898, 2013/11
Times Cited Count:42 Percentile:86.61(Instruments & Instrumentation)Flowers of the butterfly pea ( ) accumulate a group of polyacylated anthocyanins, named ternatins, in their petals. The first step in ternatin biosynthesis is the transfer of glucose from UDP-glucose to anthocyanidins such as delphinidin, a reaction catalyzed in
) accumulate a group of polyacylated anthocyanins, named ternatins, in their petals. The first step in ternatin biosynthesis is the transfer of glucose from UDP-glucose to anthocyanidins such as delphinidin, a reaction catalyzed in  by UDP-glucose:anthocyanidin 3-
by UDP-glucose:anthocyanidin 3- -glucosyltransferase (
-glucosyltransferase ( 3GT-A; AB185904). To elucidate the structure-function relationship of
3GT-A; AB185904). To elucidate the structure-function relationship of  3GT-A, recombinant
3GT-A, recombinant  3GT-A was expressed in
3GT-A was expressed in 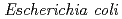 and its tertiary structure was determined to 1.85
and its tertiary structure was determined to 1.85  , resolution by using X-ray crystallography. The structure of
, resolution by using X-ray crystallography. The structure of  3GT-A shows a common folding topology, the GT-B fold, comprised of two Rossmann-like
3GT-A shows a common folding topology, the GT-B fold, comprised of two Rossmann-like  /
/ /
/ domains and a cleft located between the N- and C-domains containing two cavities that are used as binding sites for the donor (UDP-Glc) and acceptor substrates. By comparing the structure of
domains and a cleft located between the N- and C-domains containing two cavities that are used as binding sites for the donor (UDP-Glc) and acceptor substrates. By comparing the structure of  3GT-A with that of the flavonoid glycosyltransferase
3GT-A with that of the flavonoid glycosyltransferase 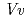 GT1 from red grape (
GT1 from red grape (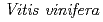 ) in complex with UDP-2-deoxy-2-fluoro glucose and kaempferol, locations of the catalytic His-Asp dyad and the residues involved in recognizing UDP-2-deoxy-2-fluoro glucose were essentially identical in
) in complex with UDP-2-deoxy-2-fluoro glucose and kaempferol, locations of the catalytic His-Asp dyad and the residues involved in recognizing UDP-2-deoxy-2-fluoro glucose were essentially identical in  3GT-A, but certain residues of
3GT-A, but certain residues of 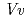 GT1 involved in binding kaempferol were found to be substituted in
GT1 involved in binding kaempferol were found to be substituted in  3GT-A. These findings are important for understanding the differentiation of acceptor-substrate recognition in these two enzymes.
3GT-A. These findings are important for understanding the differentiation of acceptor-substrate recognition in these two enzymes.
Kurihara, Kazuo*; Hirano, Yu*; Hiromoto, Takeshi*; Tamura, Itaru; Tamada, Taro*
no journal, ,
no abstracts in English
 - glucosyltransferase from
- glucosyltransferase from 
Hiromoto, Takeshi; Honjo, Eijiro*; Tamada, Taro; Kuroki, Ryota
no journal, ,
Shibazaki, Chie; Adachi, Motoyasu; Hiromoto, Takeshi; Shimizu, Rumi; Kuroki, Ryota
no journal, ,
Casein kinase 2 (CK2) is one of the ubiquitous Ser/Thr kinases and is involved in the cell cycle and the survival and proliferation of cells. CK2 is a heterotetrameric structure comprising two  - or
- or  -subunits and two regulatory
-subunits and two regulatory  -subunits. In order to understand the biological function of the alpha catalytic subunit of CK2
-subunits. In order to understand the biological function of the alpha catalytic subunit of CK2 , we aim to analyze the structure of CK2
, we aim to analyze the structure of CK2 including information of the hydrogen and hydrating water molecule by neutron crystallography. The gene coding CK2
including information of the hydrogen and hydrating water molecule by neutron crystallography. The gene coding CK2 was inserted into pET24a and expressed in E. coli strain BL21DE3, in which the mobile region and chemically reactive thiols were removed by amino acid mutation. A total of 150 mg protein was obtained from a 6 L culture, and was used for crystallization trials. The preparation of large crystals was performed using a macro seeding method specially developed for CK2
was inserted into pET24a and expressed in E. coli strain BL21DE3, in which the mobile region and chemically reactive thiols were removed by amino acid mutation. A total of 150 mg protein was obtained from a 6 L culture, and was used for crystallization trials. The preparation of large crystals was performed using a macro seeding method specially developed for CK2 . Finally, a large crystal with a volume of approximately 2 mm
. Finally, a large crystal with a volume of approximately 2 mm was reproducibly obtained. From the X-ray diffraction study, we confirmed that the crystals obtained diffracted to approx. 1
was reproducibly obtained. From the X-ray diffraction study, we confirmed that the crystals obtained diffracted to approx. 1  resolution at 100 K after soaking the crystal into the deuterated cryo protectant. The neutron diffraction data collection is planned to obtain a high resolution neutron structure of CK2
resolution at 100 K after soaking the crystal into the deuterated cryo protectant. The neutron diffraction data collection is planned to obtain a high resolution neutron structure of CK2 .
.
 -glucosyltransferase from
-glucosyltransferase from 
Hiromoto, Takeshi; Honjo, Eijiro*; Tamada, Taro; Kuroki, Ryota; Noda, Hisanobu*; Kazuma, Kohei*; Suzuki, Masahiko*
no journal, ,
UDP-glucose: anthocyanidin 3- -glucosyltransferase from
-glucosyltransferase from  (
( 3GT-A) catalyzes the transfer of glucose from UDP-glucose to anthocyanidins such as delphinidin. The glucosylation of delphinidin at the 3-hydroxyl group has been proposed as an initial glucosylation step toward the biosynthesis of ternatins, which are blue anthocyanins found in the petals of
3GT-A) catalyzes the transfer of glucose from UDP-glucose to anthocyanidins such as delphinidin. The glucosylation of delphinidin at the 3-hydroxyl group has been proposed as an initial glucosylation step toward the biosynthesis of ternatins, which are blue anthocyanins found in the petals of  . Although the crystal structures of several flavonoid glycosyltransferases (UGTs) were determined, the acceptor-substrate complexes were limited to the flavonol-bound forms. Here, in order to understand the acceptor-recognition scheme of
. Although the crystal structures of several flavonoid glycosyltransferases (UGTs) were determined, the acceptor-substrate complexes were limited to the flavonol-bound forms. Here, in order to understand the acceptor-recognition scheme of  3GT-A, the crystal structures in complex with anthocyanidin delphinidin and petunidin, and flavonol kaempferol were determined to resolutions of 2.6
3GT-A, the crystal structures in complex with anthocyanidin delphinidin and petunidin, and flavonol kaempferol were determined to resolutions of 2.6  , 2.7
, 2.7  , and 1.8
, and 1.8  respectively. The enzyme recognition of unstable anthocyanidins was firstly observed in this structural determination; nevertheless, the molecular orientations of these three acceptors in the binding site are different from those of the known flavonoid UGTs,
respectively. The enzyme recognition of unstable anthocyanidins was firstly observed in this structural determination; nevertheless, the molecular orientations of these three acceptors in the binding site are different from those of the known flavonoid UGTs, 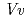 GT1 and UGT78G1. The crystal structures of
GT1 and UGT78G1. The crystal structures of  3GT-A provide insight not only into anthocyanidin configurations in enzyme, but also into a different binding scheme for acceptor-substrate recognition compared with the known UGTs.
3GT-A provide insight not only into anthocyanidin configurations in enzyme, but also into a different binding scheme for acceptor-substrate recognition compared with the known UGTs.
Hiromoto, Takeshi; Shimizu, Rumi; Adachi, Motoyasu; Shibazaki, Chie; Kuroki, Ryota
no journal, ,
no abstracts in English
Kurihara, Kazuo*; Tamura, Itaru; Hirano, Yu*; Hiromoto, Takeshi*; Tamada, Taro*
no journal, ,
no abstracts in English
Kurihara, Kazuo*; Tamura, Itaru; Hirano, Yu*; Hiromoto, Takeshi*; Kono, Fumiaki*; Tamada, Taro*
no journal, ,
Kurihara, Kazuo*; Hirano, Yu*; Hiromoto, Takeshi*; Kono, Fumiaki*; Tamura, Itaru; Tamada, Taro*
no journal, ,
no abstracts in English
Kurihara, Kazuo*; Hirano, Yu*; Hiromoto, Takeshi*; Kono, Fumiaki*; Tamura, Itaru; Tamada, Taro*
no journal, ,
no abstracts in English
Hiromoto, Takeshi; Adachi, Motoyasu; Shibazaki, Chie; Kuroki, Ryota
no journal, ,
T4 phage lysozyme (T4L) is an endoacetylmuramidase that degrades the murein of the bacterial cell wall by cleavage of the  -1,4-glycosidic bond between
-1,4-glycosidic bond between 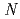 -acetylmuramic acid and
-acetylmuramic acid and 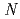 -acetylglucosamine. We previously reported that the substitution of the catalytic Thr26 to the nucleophilic His converts the wild type (WT) T4L from an inverting to a retaining glycosidase, in which the
-acetylglucosamine. We previously reported that the substitution of the catalytic Thr26 to the nucleophilic His converts the wild type (WT) T4L from an inverting to a retaining glycosidase, in which the  -configuration of the substrate is retained in the product. It was also found that the Thr26His mutant T4L can catalyze the transglycosylation reaction more effectively than hydrolysis although the WT T4L has no transglycosidase activity. To clarify the role of the substituted His26 on transglycosylation and its relationship to the neighboring acidic residue Asp20 by neutron crystallography, the perdeuterated recombinant proteins of the WT and Thr26His mutant T4L were prepared for crystallization in this study. The perdeuterated forms were produced in
-configuration of the substrate is retained in the product. It was also found that the Thr26His mutant T4L can catalyze the transglycosylation reaction more effectively than hydrolysis although the WT T4L has no transglycosidase activity. To clarify the role of the substituted His26 on transglycosylation and its relationship to the neighboring acidic residue Asp20 by neutron crystallography, the perdeuterated recombinant proteins of the WT and Thr26His mutant T4L were prepared for crystallization in this study. The perdeuterated forms were produced in 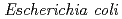 cells cultured in deuterated rich media. After purification, macroseeding was performed to grow large crystals by transferring individual crystals to hanging drops. A crystal of Thr26His mutant T4L with a volume of 0.1 mm
cells cultured in deuterated rich media. After purification, macroseeding was performed to grow large crystals by transferring individual crystals to hanging drops. A crystal of Thr26His mutant T4L with a volume of 0.1 mm was grown after one month. Preliminary neutron-diffraction experiment at the research reactor FRM-II (Munich, Germany) at 100 K gave diffraction spots beyond 2.5
was grown after one month. Preliminary neutron-diffraction experiment at the research reactor FRM-II (Munich, Germany) at 100 K gave diffraction spots beyond 2.5  resolution for 1.5 hour exposure.
resolution for 1.5 hour exposure.
Shimizu, Rumi; Hiromoto, Takeshi; Adachi, Motoyasu; Shibazaki, Chie; Kuroki, Ryota
no journal, ,
no abstracts in English
Shimizu, Rumi; Hiromoto, Takeshi; Adachi, Motoyasu; Kuroki, Ryota; Kataoka, Misumi*; Ishikawa, Kazuhiko*
no journal, ,
For neutron crystal structure analysis of proteins, it is necessary to prepare large crystals in volume (several mm ) compared to that required for X-ray crystal structure analysis. To prepare the large volume crystal, we inevitably need much amount of purified protein. As a development in preparation of samples for neutron crystal structure analysis of proteins, we have tried to develop expression system for many kinds of protein using Eschericha coli, Brevibacillus, Pichia pastoris, cultivation cell and so on. Recently, we have succeeded in high level expression of cellulose (EGPf) derived from Archaea
) compared to that required for X-ray crystal structure analysis. To prepare the large volume crystal, we inevitably need much amount of purified protein. As a development in preparation of samples for neutron crystal structure analysis of proteins, we have tried to develop expression system for many kinds of protein using Eschericha coli, Brevibacillus, Pichia pastoris, cultivation cell and so on. Recently, we have succeeded in high level expression of cellulose (EGPf) derived from Archaea 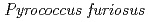 using Brevibacillus system.
using Brevibacillus system.
Shibazaki, Chie; Adachi, Motoyasu; Hiromoto, Takeshi; Shimizu, Rumi; Kuroki, Ryota
no journal, ,
Casein kinase 2(CK2) is one of the ubiquitous Ser/Thr kinases and is involved in the cell cycle and the survival and proliferation of cells. In order to understand the biological function of the alpha catalytic subunit of CK2 (CK2a), we aim to analyze the structure of CK2a including information of the hydrogen and hydrating water molecule by neutron crystallography. The gene coding CK2a was expressed in E. coli, in which the mobile region and chemically reactive thiols were removed by amino acid mutation. The preparation of large crystals with inhibitor (Emodin and CX4945) was performed using a macro seeding method. Finally, large crystals with a volume of approximately 2 mm were reproducibly obtained. The crystals were dialyzed in the deuterated reagent and deuterium water. We have collected high resolution neutron diffraction images of emodin complex and CX-4945 complex at neutron beam line BioDIFF (FRM-II, Munich).
were reproducibly obtained. The crystals were dialyzed in the deuterated reagent and deuterium water. We have collected high resolution neutron diffraction images of emodin complex and CX-4945 complex at neutron beam line BioDIFF (FRM-II, Munich).
Kurihara, Kazuo*; Hirano, Yu*; Hiromoto, Takeshi*; Tamura, Itaru; Tamada, Taro*
no journal, ,
no abstracts in English