Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Papadopoulos, A.*; Kyriakou, I.*; Matsuya, Yusuke; Incerti, S.*; Daglis, I. A.*; Emfietzoglou, D.*
Applied Sciences (Internet), 12(18), p.8950_1 - 8950_20, 2022/09
Times Cited Count:2 Percentile:28.33(Chemistry, Multidisciplinary)The quality factor (Q) is the index which is used when evaluating the stochastic (e.g., carcinogenic) risk of diverse ionizing radiations. While the Q value can be commonly determined from the Linear Energy Transfer (LET), more elaborate approaches are based on the microdosimetric parameter lineal energy (y) calculated either by analytical model or Monte Carlo (MC) simulations. However, the developing of the model for determining the Q value is still ongoing worldwide for realizing the precise risk assessment. In this study, various generalized analytical models that account for both 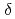 -ray transport and energy-loss straggling effects are utilized to evaluate the Q values over the proton energy range 1-250 MeV. The results revealed that the LET-based ICRP Report 60 recommendations underestimate the microdosimetric-based Q values of protons with energy below 100 MeV, which was also confirmed by using MC simulation data on the y values. The present work shows that analytic models may offer a practical alternative to computer-intensive MC simulations for calculating Q values based on the microdosimetric methodologies. In future study, we are planning to compare the y spectra and subsequent calculations of Q based on new MC data with the latest versions of Geant4-DNA and PHITS track structure codes which make use of different physics models.
-ray transport and energy-loss straggling effects are utilized to evaluate the Q values over the proton energy range 1-250 MeV. The results revealed that the LET-based ICRP Report 60 recommendations underestimate the microdosimetric-based Q values of protons with energy below 100 MeV, which was also confirmed by using MC simulation data on the y values. The present work shows that analytic models may offer a practical alternative to computer-intensive MC simulations for calculating Q values based on the microdosimetric methodologies. In future study, we are planning to compare the y spectra and subsequent calculations of Q based on new MC data with the latest versions of Geant4-DNA and PHITS track structure codes which make use of different physics models.
Schuemann, J.*; McNamara, A. L.*; Warmenhoven, J. W.*; Henthorn, N. T.*; Kirkby, K.*; Merchant, M. J.*; Ingram, S.*; Paganetti, H.*; Held, K. D.*; Ramos-Mendez, J.*; et al.
Radiation Research, 191(1), p.76 - 93, 2019/01
Times Cited Count:47 Percentile:94.65(Biology)We propose a new Standard DNA Damage (SDD) data format to unify the interface between the simulation of damage induction in DNA and the biological modelling of DNA repair processes, and introduce the effect of the environment (molecular oxygen or other compounds) as a flexible parameter. Such a standard greatly facilitates inter-model comparisons, providing an ideal environment to tease out model assumptions and identify persistent, underlying mechanisms. Through inter-model comparisons, this unified standard has the potential to greatly advance our understanding of the underlying mechanisms of radiation-induced DNA damage and the resulting observable biological effects when radiation parameters and/or environmental conditions change.