Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Inoue, Rintaro*; Oda, Takashi; Nakagawa, Hiroshi; Tominaga, Taiki*; Ikegami, Takahisa*; Konuma, Tsuyoshi*; Iwase, Hiroki*; Kawakita, Yukinobu; Sato, Mamoru*; Sugiyama, Masaaki*
Biophysical Journal, 124(3), p.540 - 548, 2025/02
Times Cited Count:0 Percentile:0.00(Biophysics)Ueji, Rintaro*; Gong, W.; Harjo, S.; Kawasaki, Takuro; Shibata, Akinobu*; Kimura, Yuji*; Inoue, Tadanobu*; Tsuchida, Noriyuki*
ISIJ International, 64(2), p.459 - 465, 2024/01
Times Cited Count:1 Percentile:41.92(Metallurgy & Metallurgical Engineering)Tominaga, Taiki*; Nakagawa, Hiroshi; Sahara, Masae*; Oda, Takashi*; Inoue, Rintaro*; Sugiyama, Masaaki*
Life (Internet), 12(5), p.675_1 - 675_9, 2022/05
Times Cited Count:2 Percentile:21.16(Biology)The background scattering of sample cells suitable for aqueous protein solution samples, conducted with a neutron backscattering spectrometer, was evaluated. It was found that the scattering intensity of an aluminum sample cell coated with boehmite using D O was lower than that of a sample cell coated with regular water (H
O was lower than that of a sample cell coated with regular water (H O). In addition, meticulous attention to cells with small individual weight differences and the positional reproducibility of the sample cell relative to the spectrometer neutron beam position enabled the accurate subtraction of the scattering profiles of the D
O). In addition, meticulous attention to cells with small individual weight differences and the positional reproducibility of the sample cell relative to the spectrometer neutron beam position enabled the accurate subtraction of the scattering profiles of the D O buffer and the sample container. Consequently, high quality information on protein dynamics could be extracted from dilute protein solutions.
O buffer and the sample container. Consequently, high quality information on protein dynamics could be extracted from dilute protein solutions.
Nakagawa, Hiroshi; Saio, Tomohide*; Nagao, Michihiro*; Inoue, Rintaro*; Sugiyama, Masaaki*; Ajito, Satoshi; Tominaga, Taiki*; Kawakita, Yukinobu
Biophysical Journal, 120(16), p.3341 - 3354, 2021/08
Times Cited Count:7 Percentile:44.90(Biophysics)A multi-domain protein can have various conformations in solution. Interactions with other molecules result in the stabilization of one of the conformations and change in the domain dynamics. SAXS, a well-established experimental technique, can be employed to elucidate the conformation of a multi-domain protein in solution. NSE spectroscopy is a promising technique for recording the domain dynamics in nanosecond and nanometer scale. Despite the great efforts, there are still under development. Thus, we quantitatively removed the contribution of diffusion dynamics and hydrodynamic interactions from the NSE data via incoherent scattering, revealing the differences in the domain dynamics of the three functional states of a multi-domain protein, MurD. The differences among the three states can be explained by two domain modes.
Inoue, Rintaro*; Oda, Takashi*; Nakagawa, Hiroshi; Tominaga, Taiki*; Saio, Tomohide*; Kawakita, Yukinobu; Shimizu, Masahiro*; Okuda, Aya*; Morishima, Ken*; Sato, Nobuhiro*; et al.
Scientific Reports (Internet), 10, p.21678_1 - 21678_10, 2020/12
Times Cited Count:8 Percentile:26.35(Multidisciplinary Sciences)Incoherent quasielastic neutron scattering (iQENS) is a fascinating technique for investigating the internal dynamics of protein. However, both low flux of neutron beam and absence of analytical procedure for extracting the internal dynamics from iQENS profile have been obstacles for studying it under physiological condition (in solution). Thanks to the recent development of neutron source, spectrometer and computational technique, they enable us to decouple internal dynamics, translational and rotational diffusions from the iQENS profile. The internal dynamics of two proteins: globular domain protein (GDP) and intrinsically disordered protein (IDP) in solution were studied. It was found that the average relaxation rate of IDP was larger than that of GDP. Through the detailed analyses on their internal dynamics, it was revealed that the fraction of mobile H atoms in IDP was much higher than that in GDP. Interestingly, the fraction of mobile H atoms was closely related to the fraction of H atoms on highly solvent exposed surfaces. The iQENS study presented that the internal dynamics were governed by the highly solvent exposed amino acid residues depending upon protein molecular architectures.
 -olefin using small-angle X-ray scattering
-olefin using small-angle X-ray scatteringOba, Yojiro; Motokawa, Ryuhei; Hino, Masahiro*; Adachi, Nozomu*; Todaka, Yoshikazu*; Inoue, Rintaro*; Sugiyama, Masaaki*
Chemistry Letters, 49(7), p.823 - 825, 2020/07
Times Cited Count:0 Percentile:0.00(Chemistry, Multidisciplinary)Sugiyama, Masaaki*; Inoue, Rintaro*; Nakagawa, Hiroshi; Saio, Tomohide*
Hamon, 30(1), p.16 - 25, 2020/02
Neutron has distinct features as a scattering probe to analyze structure and dynamics of biological macromolecules. The theme of this review is to try to describe how we did/do utilize them. And "How we should utilize them more effectively in the trend of integrative structural biology?" with solution scattering.
Inoue, Rintaro*; Kanaya, Toshiji*; Yamada, Takeshi*; Shibata, Kaoru; Fukao, Koji*
Physical Review E, 97(1), p.012501_1 - 012501_6, 2018/01
Times Cited Count:10 Percentile:60.88(Physics, Fluids & Plasmas)In this study, we investigate the  process of a polystyrene thin film using inelastic neutron scattering (INS), dielectric relaxation spectroscopy (DRS), and thermal expansion spectroscopy (TES). The DRS and TES measurements exhibited a decrease in glass transition temperature (
process of a polystyrene thin film using inelastic neutron scattering (INS), dielectric relaxation spectroscopy (DRS), and thermal expansion spectroscopy (TES). The DRS and TES measurements exhibited a decrease in glass transition temperature (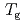 ) with film thickness. On the other hand, an increase in
) with film thickness. On the other hand, an increase in 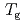 was observed in INS studies. In order to interpret this contradiction, we investigated the temperature dependence of the peak frequency (
was observed in INS studies. In order to interpret this contradiction, we investigated the temperature dependence of the peak frequency (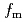 ) of the
) of the  process probed by DRS and TES. The experiments revealed an increase in the peak frequency (
process probed by DRS and TES. The experiments revealed an increase in the peak frequency (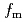 ) with decreasing film thickness in the frequency region. This observation is consistent with the observed decrease in
) with decreasing film thickness in the frequency region. This observation is consistent with the observed decrease in 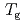 with thickness. The discrepancy between INS and DRS or TES descriptions of the
with thickness. The discrepancy between INS and DRS or TES descriptions of the  process is likely to be attributed to a decrease in the apparent activation energy with film thickness and reduced mobility, due to the impenetrable wall effect.
process is likely to be attributed to a decrease in the apparent activation energy with film thickness and reduced mobility, due to the impenetrable wall effect.
Sugiyama, Masaaki*; Nakagawa, Hiroshi; Inoue, Rintaro*; Kawakita, Yukinobu
JAEA-Review 2017-024, 40 Pages, 2017/12
Now-a-days, promotion of life science by utilizing neutron (neutrons biology) is highly demanded in our country, following installation and improvement of high quality and intensity neutron sources at J-PARC and JRR-3. Aiming at accelerating development of neutrons biology in our country, an international workshop "Neutron biology for next generation" was held as a J-PARC Workshop at Ibaraki Quantum Beam Research Center from 22 March to 23 March in 2017. In the workshop, latest instruments, new-fashioned methodologies, recent scientific results and future perspectives were extensively discussed by domestic neutron instrumental scientists and domestic/foreign neutron biologists. This is a report of the workshop summarized by organizers.
Oba, Yojiro*; Morooka, Satoshi; Oishi, Kazuki*; Suzuki, Junichi*; Takata, Shinichi; Sato, Nobuhiro*; Inoue, Rintaro*; Tsuchiyama, Toshihiro*; Gilbert, E. P.*; Sugiyama, Masaaki*
Journal of Applied Crystallography, 50(2), p.334 - 339, 2017/04
Times Cited Count:3 Percentile:27.09(Chemistry, Multidisciplinary)Oba, Yojiro*; Morooka, Satoshi; Sato, Hirotaka*; Sato, Nobuhiro*; Inoue, Rintaro*; Sugiyama, Masaaki*
Hamon, 26(4), p.170 - 173, 2016/11
Oba, Yojiro*; Morooka, Satoshi; Oishi, Kazuki*; Sato, Nobuhiro*; Inoue, Rintaro*; Adachi, Nozomu*; Suzuki, Junichi*; Tsuchiyama, Toshihiro*; Gilbert, E. P.*; Sugiyama, Masaaki*
Journal of Applied Crystallography, 49(5), p.1659 - 1664, 2016/10
Times Cited Count:13 Percentile:63.41(Chemistry, Multidisciplinary)Endo, Hitoshi; Sugiyama, Masaaki*; Inoue, Rintaro*
Hamon, 22(3), p.258 - 267, 2012/08
In this text, the authors introduce two powerful techniques and one developing one for Small-Angle Neutron Scattering (SANS). The most fascinating feature of neutron as a scattering probe is its isotope effect in hydrogen. The first topic is concerning about recent progress on Contrast Variation Method: the author shows how to apply the contrast variation method to protein-mineral complex system and analyze the data. The second is concerning about deuteration-labeling: the author shows kinetics analysis in quaternary structure of homo-oligomeric protein with this technique. The final topic is concerning about the next generation analysis: an analysis method coupling SANS with neutron spin echo for dynamics of tertiary structure of protein.
Takahashi, Nobuaki; Nishida, Koji*; Inoue, Rintaro*; Ogawa, Hiroki*; Kanaya, Toshiji*; Nagao, Michihiro*
NSL News Letter, 2007-4, p.155 - 157, 2007/04
We have studied dynamics of poly(vinyl alcohol) (PVA) gel in a mixture of deuterated dimethyl sulfoxide (DMSO-d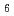 ) and D
) and D O (60/40 by volume) during heating process from 25
O (60/40 by volume) during heating process from 25 C to 80
C to 80 C using neutron spin-echo (NSE) techniques.
C using neutron spin-echo (NSE) techniques.
Kanaya, Toshiji; Takahashi, Nobuaki; Inoue, Rintaro*; Matsuba, Go*; Nishida, Koji*; Nagao, Michihiro*
ISSP Activity Report on Neutron scattering Research; Experimental Reports (CD-ROM), 13, 1 Pages, 2006/00
Takahashi, Nobuaki; Nishida, Koji*; Tsubouchi, Tsuyoshi*; Ogawa, Hiroki*; Inoue, Rintaro*; Kanaya, Toshiji*; Nagao, Michihiro*
ISSP Activity Report on Neutron scattering Research; Experimental Reports (CD-ROM), 13, 2 Pages, 2006/00
no abstracts in English
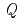 range
rangeKanaya, Toshiji*; Kakurai, Kazuhisa; Tsukushi, Itaru*; Inoue, Rintaro*; Watanabe, Hiroshi*; Nishi, Masakazu*; Nakajima, Kenji; Takemura, Kazuhiro*; Furuya, Hidemine*
Journal of the Physical Society of Japan, 74(12), p.3236 - 3240, 2005/12
Times Cited Count:6 Percentile:41.10(Physics, Multidisciplinary)We performed thermal neutron spin echo measurements on a glass-forming polymer, deuterated polybutadiene (-[CD -CD=CD-CD
-CD=CD-CD ]
]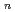 -; PB-d
-; PB-d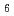 ), in a high
), in a high 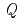 range up to 3.5
range up to 3.5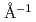 covering the first and second peaks of the structure factor S(
covering the first and second peaks of the structure factor S(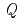 ), and evaluated the decay rate
), and evaluated the decay rate 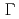 (
(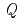 ) of the coherent intermediate scattering function as a function of
) of the coherent intermediate scattering function as a function of 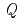 . In contrast to the previous experimental findings on the first peak of S(
. In contrast to the previous experimental findings on the first peak of S(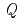 ) at
) at 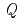 = 1.5
= 1.5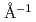 , no "de Gennes" type narrowing was observed on the second peak at
, no "de Gennes" type narrowing was observed on the second peak at 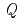 = 2.9
= 2.9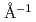 . This novel finding indicates that the narrowing is hidden by the distinct motions of the soft -CD
. This novel finding indicates that the narrowing is hidden by the distinct motions of the soft -CD -CD
-CD - and =CD-CD
- and =CD-CD - units and the self-motions of the rigid and soft units in the
- units and the self-motions of the rigid and soft units in the 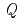 range around the second peak.
range around the second peak.
Nakagawa, Hiroshi; Saio, Tomohide*; Oda, Takashi*; Sato, Mamoru*; Inoue, Rintaro*; Sugiyama, Masaaki*; Tominaga, Taiki*; Kawakita, Yukinobu
no journal, ,
Protein is thermally fluctuating in solution, and the dynamics is essential for its biological functions. A protein has hierarchal structure and dynamics in temporal and spatial scale. In this work, QENS of a multi-domain protein, MurD, were measured by the TOF near backscattering spectrometer DNA in order to observe the internal motions. Hef is classified as an intrinsically disordered protein, which lost the rigid folded domain structure, and then have a more flexible structure than a folded protein. We will discuss the data treatment and analytical method of QENS of protein solutions, and characteristic dynamical features of folded rigid and disordered flexible proteins in the presentation.
Nakagawa, Hiroshi; Saio, Tomohide*; Nagao, Michihiro*; Inoue, Rintaro*; Sugiyama, Masaaki*; Tominaga, Taiki*; Kawakita, Yukinobu
no journal, ,
The flexible conformation of a multidomain protein is responsible for its biological function. 3-domain protein: MurD (47kDa) changes its domain conformation sequentially from open to semi-closed to closed conformation during enzymatic reactions. However, the dynamics of the domains in each conformation is unknown. In this study, we combined small-angle X-ray and neutron scattering (SAXS and SANS), dynamic light scattering (DLS), neutron back scattering (NBS), neutron spin echo (NSE), and molecular dynamics (MD) simulations to investigate the conformational dynamics of MurD in the three corresponding states (apo and ATP, inhibitor-bound state). The analysis showed that the conformational dynamics of MurD during the enzymatic reaction was not affected. The analysis suggests that the changes in domain dynamics during enzymatic reactions are related to the affinity and reaction efficiency with ligands that bind specifically to each reaction state.
Nakagawa, Hiroshi; Saio, Tomohide*; Nagao, Michihiro*; Inoue, Rintaro*; Sugiyama, Masaaki*; Tominaga, Taiki*; Kawakita, Yukinobu
no journal, ,
The flexible conformation of a multidomain protein is responsible for its biological function. 3-domain protein: MurD (47kDa) changes its domain conformation sequentially from open to semi-closed to closed conformation during enzymatic reactions. However, the dynamics of the domains in each conformation is unknown. In this study, we combined small-angle X-ray and neutron scattering (SAXS and SANS), dynamic light scattering (DLS), neutron back scattering (NBS), neutron spin echo (NSE), and molecular dynamics (MD) simulations to investigate the conformational dynamics of MurD in the three corresponding states (apo and ATP, inhibitor-bound state). The analysis showed that the conformational dynamics of MurD during the enzymatic reaction was not affected. The analysis suggests that the changes in domain dynamics during enzymatic reactions are related to the affinity and reaction efficiency with ligands that bind specifically to each reaction state.