Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
 binding site of a
binding site of a  -lactamase from the halophile by anomalous X-ray diffraction
-lactamase from the halophile by anomalous X-ray diffractionArai, Shigeki; Shibazaki, Chie; Shimizu, Rumi; Adachi, Motoyasu; Tamada, Taro; Tokunaga, Hiroko*; Ishibashi, Matsujiro*; Tokunaga, Masao*; Kuroki, Ryota
Kyushu Shinkurotoronko Kenkyu Senta Nempo, 2014, p.17 - 19, 2016/03
no abstracts in English
 sp. AS-131 isolated from Antarctic Ocean
sp. AS-131 isolated from Antarctic OceanYonezawa, Yasushi*; Nagayama, Aiko*; Tokunaga, Hiroko*; Ishibashi, Matsujiro*; Arai, Shigeki; Kuroki, Ryota; Watanabe, Keiichi*; Arakawa, Tsutomu*; Tokunaga, Masao*
Protein Journal, 34(4), p.275 - 283, 2015/08
Times Cited Count:4 Percentile:10.01(Biochemistry & Molecular Biology)Nucleoside diphosphate kinase isolated from psychrophilic  sp. AS-131 (ASNDK) was expressed in
sp. AS-131 (ASNDK) was expressed in  and purified to homogeneity. Comparing to mesophilic NDK isolated from
and purified to homogeneity. Comparing to mesophilic NDK isolated from  , ASNDK exhibited highly elevated thermolability: (1)
, ASNDK exhibited highly elevated thermolability: (1) 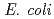 expression at 37
expression at 37 C as a denatured insoluble form, and (2) 30
C as a denatured insoluble form, and (2) 30 C lower optimum temperature of enzymatic activity. The subunit structure of ASNDK was suggested to be dimer, as in NDKs isolated from moderate halophiles.
C lower optimum temperature of enzymatic activity. The subunit structure of ASNDK was suggested to be dimer, as in NDKs isolated from moderate halophiles.
 -lactamase from the moderate halophile
-lactamase from the moderate halophile  sp.560 and the discovery of a Cs
sp.560 and the discovery of a Cs -selective binding site
-selective binding siteArai, Shigeki; Yonezawa, Yasushi*; Okazaki, Nobuo*; Matsumoto, Fumiko*; Shibazaki, Chie; Shimizu, Rumi; Yamada, Mitsugu*; Adachi, Motoyasu; Tamada, Taro; Kawamoto, Masahide*; et al.
Acta Crystallographica Section D, 71(3), p.541 - 554, 2015/03
Times Cited Count:8 Percentile:52.61(Biochemical Research Methods)The crystal structure of halophilic  -lactamase from
-lactamase from  sp.560 (HaBLA) was determined using X-ray crystallography. Moreover, the locations of bound Sr
sp.560 (HaBLA) was determined using X-ray crystallography. Moreover, the locations of bound Sr and Cs
and Cs ions were identified by anomalous X-ray diffraction. The location of one Cs
ions were identified by anomalous X-ray diffraction. The location of one Cs specific binding site was identified on HaBLA even in the presence of 9-fold molar excess of Na
specific binding site was identified on HaBLA even in the presence of 9-fold molar excess of Na (90 mM Na
(90 mM Na /10 mM Cs
/10 mM Cs ). This Cs
). This Cs binding site is formed by two main-chain O atoms and an aromatic ring of a side chain of Trp. An aromatic ring of Trp interacts with Cs
binding site is formed by two main-chain O atoms and an aromatic ring of a side chain of Trp. An aromatic ring of Trp interacts with Cs by the cation-
by the cation-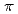 interaction. The observation of a selective and high-affinity Cs
interaction. The observation of a selective and high-affinity Cs binding site provides important information that is useful for designing artificial Cs
binding site provides important information that is useful for designing artificial Cs binding sites useful in bioremediation of radioactive isotopes.
binding sites useful in bioremediation of radioactive isotopes.
 sp.593
sp.593Arai, Shigeki; Yonezawa, Yasushi*; Ishibashi, Matsujiro*; Matsumoto, Fumiko*; Adachi, Motoyasu; Tamada, Taro; Tokunaga, Hiroko*; Blaber, M.; Tokunaga, Masao*; Kuroki, Ryota
Acta Crystallographica Section D, 70(3), p.811 - 820, 2014/03
Times Cited Count:13 Percentile:63.53(Biochemical Research Methods)In order to clarify the structural basis of halophilic characteristics of an alkaline phosphatase derived from the moderate halophile  sp.593 (HaAP), the tertiary structure of HaAP was determined to 2.1
sp.593 (HaAP), the tertiary structure of HaAP was determined to 2.1 resolution by X-ray crystallography. Structural properties of surface negative charge and core hydrophobicity are shown to be intermediate between halophile and non-halophile characteristics, and may explain the unique functional adaptation to a wide-range of salt concentration.
resolution by X-ray crystallography. Structural properties of surface negative charge and core hydrophobicity are shown to be intermediate between halophile and non-halophile characteristics, and may explain the unique functional adaptation to a wide-range of salt concentration.
Ishibashi, Matsujiro*; Uchino, Manami*; Arai, Shigeki; Kuroki, Ryota; Arakawa, Tsutomu*; Tokunaga, Masao*
Archives of Biochemistry and Biophysics, 525(1), p.47 - 52, 2012/09
Times Cited Count:7 Percentile:19.87(Biochemistry & Molecular Biology)Nucleoside diphosphate kinase (HsNDK) from Halobacterium salinarum requires salt at high concentrations for folding. A D148C mutant, in which Asp148 was replaced with Cys, was designed to enhance stability and folding in low salt solution by S-S bond. It showed increased thermal stability by about 10 C in 0.2 M NaCl over the wild type HsNDK. It refolded from heat-denaturation even in 0.1 M NaCl, while the wild type required 2 M NaCl to achieve the same level of activity recovery. This enhanced refolding is due to the three S-S bonds between two basic dimeric units in the hexameric HsNDK structure. Moreover, salt concentration dependency of heat-denaturation process and refolding process of the wild type and D148C mutant HsNDKs were investigated by the circular dichroism and native-PAGE analysis.
C in 0.2 M NaCl over the wild type HsNDK. It refolded from heat-denaturation even in 0.1 M NaCl, while the wild type required 2 M NaCl to achieve the same level of activity recovery. This enhanced refolding is due to the three S-S bonds between two basic dimeric units in the hexameric HsNDK structure. Moreover, salt concentration dependency of heat-denaturation process and refolding process of the wild type and D148C mutant HsNDKs were investigated by the circular dichroism and native-PAGE analysis.
 sp. nucleoside diphosphate kinase
sp. nucleoside diphosphate kinaseArai, Shigeki; Yonezawa, Yasushi; Okazaki, Nobuo; Matsumoto, Fumiko; Tamada, Taro; Tokunaga, Hiroko*; Ishibashi, Matsujiro*; Blaber, M.; Tokunaga, Masao*; Kuroki, Ryota
Protein Science, 21(4), p.498 - 510, 2012/04
Times Cited Count:15 Percentile:34.92(Biochemistry & Molecular Biology)In order to clarify the oligomer state of nucleoside diphosphate kinase (NDK) from moderately halophilic  sp. 593 (HaNDK), the crystal structure of HaNDK was determined by X-ray crystallography. The crystal structures of the wild-type HaNDK and the mutant HaNDK (E134A) showed a dimer and a tetramer, respectively. The higher ordered association of proteins usually contributes to an increase in thermal stability and substrate affinity. The change in the assembly form by a minimum mutation may be an effective way for NDK to acquire molecular characteristics suited to various circumstances.
sp. 593 (HaNDK), the crystal structure of HaNDK was determined by X-ray crystallography. The crystal structures of the wild-type HaNDK and the mutant HaNDK (E134A) showed a dimer and a tetramer, respectively. The higher ordered association of proteins usually contributes to an increase in thermal stability and substrate affinity. The change in the assembly form by a minimum mutation may be an effective way for NDK to acquire molecular characteristics suited to various circumstances.
Tokunaga, Hiroko*; Ishibashi, Matsujiro*; Arisaka, Fimio*; Arai, Shigeki; Kuroki, Ryota; Arakawa, Tsutomu*; Tokunaga, Masao*
FEBS Letters, 582(7), p.1049 - 1054, 2008/04
Times Cited Count:17 Percentile:39.41(Biochemistry & Molecular Biology) nucleoside diphosphate kinase (HaNDK) forms a dimeric assembly and
nucleoside diphosphate kinase (HaNDK) forms a dimeric assembly and 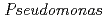 NDK (PaNDK) forms a tetrameric assembly. The mutation of Glu134 to Ala in HaNDK resulted in conversion of the native dimeric structure to the tetramer assembly. Conversely, the mutation of Ala134 to Glu in PaNDK leads to conversion from tetramer to dimer assembly, indicating that a single amino acid substitution at position 134 results in an alteration of the oligomeric structure of NDK. Modeling structure of HaNDK and PaNDK, based on the crystal structure of
NDK (PaNDK) forms a tetrameric assembly. The mutation of Glu134 to Ala in HaNDK resulted in conversion of the native dimeric structure to the tetramer assembly. Conversely, the mutation of Ala134 to Glu in PaNDK leads to conversion from tetramer to dimer assembly, indicating that a single amino acid substitution at position 134 results in an alteration of the oligomeric structure of NDK. Modeling structure of HaNDK and PaNDK, based on the crystal structure of 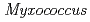 NDK, suggested sufficient repulsion by Glu134 to disrupt dimer-dimer interaction to form tetramer.
NDK, suggested sufficient repulsion by Glu134 to disrupt dimer-dimer interaction to form tetramer.
Tamada, Taro; Honjo, Eijiro; Maeda, Yoshitake*; Okamoto, Tomoyuki*; Ishibashi, Matsujiro*; Tokunaga, Masao*; Kuroki, Ryota
Proceedings of the National Academy of Sciences of the United States of America, 103(9), p.3135 - 3140, 2006/02
Times Cited Count:99 Percentile:84.91(Multidisciplinary Sciences)A crystal structure of the signaling complex between human granulocyte colony-stimulating factor (GCSF), and a ligand binding region of GCSF receptor (GCSF-R), has been determined to 2.8 resolution. The GCSF:GCSF-R complex formed a 2:2 stoichiometry via a cross-over interaction between the Ig-like domains of GCSF-R and GCSF. The conformation of the complex is quite different to that between human GCSF and the CRH domain of mouse GCSF-R, but similar to that of the interleukin-6 (IL-6)/gp130 signaling complex. The Ig-like domain cross-over structure necessary for GCSF-R activation is consistent with previously reported thermodynamic and mutational analyses.
resolution. The GCSF:GCSF-R complex formed a 2:2 stoichiometry via a cross-over interaction between the Ig-like domains of GCSF-R and GCSF. The conformation of the complex is quite different to that between human GCSF and the CRH domain of mouse GCSF-R, but similar to that of the interleukin-6 (IL-6)/gp130 signaling complex. The Ig-like domain cross-over structure necessary for GCSF-R activation is consistent with previously reported thermodynamic and mutational analyses.
Honjo, Eijiro; Tamada, Taro; Maeda, Yoshitake*; Koshiba, Takumi*; Matsukura, Yasuko*; Okamoto, Tomoyuki*; Ishibashi, Matsujiro*; Tokunaga, Masao*; Kuroki, Ryota
Acta Crystallographica Section F, 61(8), p.788 - 790, 2005/08
Times Cited Count:7 Percentile:54.65(Biochemical Research Methods)Granulocyte colony-stimulating factor (GCSF) receptor receives signals for regulating the proliferation and differentiation of the precursor cells of granulocytes. The complex composed of two GCSFs and two GCSF receptors was crystallized. The crystal of the complex was grown in 1.0 M sodium formate and 0.1 M sodium acetate (pH4.6). It belongs to the space group 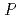 4
4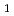 2
2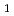 2 (or its enantiomorph
2 (or its enantiomorph 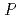 4
4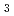 2
2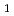 2) with unit cell dimensions of
2) with unit cell dimensions of 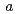 =
= 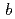 = 110.1
= 110.1  ,
, 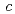 = 331.8
= 331.8  . However, the diffraction data from the crystal beyond 5
. However, the diffraction data from the crystal beyond 5  resolution could not be collected. Since the heterogeneity of GCSF receptor seems to interrupt growth of good quality crystals, the GCSF receptor was fractionated by achromatography. Crystals of GCSF/fractionated GCSF receptor complex were grown as a new crystal form in 0.2 M ammonium phosphate. The new crystal diffracts beyond 3.0
resolution could not be collected. Since the heterogeneity of GCSF receptor seems to interrupt growth of good quality crystals, the GCSF receptor was fractionated by achromatography. Crystals of GCSF/fractionated GCSF receptor complex were grown as a new crystal form in 0.2 M ammonium phosphate. The new crystal diffracts beyond 3.0  resolution and belongs to space group
resolution and belongs to space group 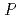 3
3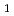 21 (or its enantiomorph
21 (or its enantiomorph 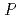 3
3 21) with unit-cell parameters
21) with unit-cell parameters 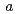 =
= 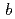 = 134.8,
= 134.8, 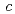 = 105.7
= 105.7  .
.
Tamada, Taro; Honjo, Eijiro; Maeda, Yoshitake*; Okamoto, Tomoyuki*; Ishibashi, Matsujiro*; Tokunaga, Masao*; Kuroki, Ryota
no journal, ,
We have succeeded in crystallization of 2:2 complex between human granulocyte colony-stimulating factor (hGCSF) and the Ig-like and CRH domains of human GCSF-R (hGCSF-R) and determined its tertiary structure by X-ray crystallography at 2.8  resolution. The signaling 2:2 complex is formed via cross-over interactions between the Ig-like domain of hGCSF-R and the neighboring hGCSF, forming a two-fold axis of crystallographic symmetry. This conformation is quite different from that of the heterogeneous mGCSF-R complex, and more closely resembles the 2:2:2 active assembly of human interleukin-6 (IL-6), human IL-6
resolution. The signaling 2:2 complex is formed via cross-over interactions between the Ig-like domain of hGCSF-R and the neighboring hGCSF, forming a two-fold axis of crystallographic symmetry. This conformation is quite different from that of the heterogeneous mGCSF-R complex, and more closely resembles the 2:2:2 active assembly of human interleukin-6 (IL-6), human IL-6  -receptor and human gp130 (which is a shared signal transducing receptor for several cytokines), and the 2:2 assembly of viral IL-6 and human gp130. The Ig-like domain cross-over structure necessary for GCSF-R activation is consistent with previously reported thermodynamic and mutational analyses.
-receptor and human gp130 (which is a shared signal transducing receptor for several cytokines), and the 2:2 assembly of viral IL-6 and human gp130. The Ig-like domain cross-over structure necessary for GCSF-R activation is consistent with previously reported thermodynamic and mutational analyses.
Tokunaga, Hiroko*; Ishibashi, Matsujiro*; Arisaka, Fimio*; Arai, Shigeki; Kuroki, Ryota; Yamaguchi, Rui*; Arakawa, Tsutomu*; Tokunaga, Masao*
no journal, ,
no abstracts in English
Arai, Shigeki; Yonezawa, Yasushi; Okazaki, Nobuo; Tamada, Taro; Tokunaga, Hiroko*; Ishibashi, Matsujiro*; Tokunaga, Masao*; Kuroki, Ryota
no journal, ,
The nucleoside diphosphate kinases (NDKs) are known to have a tetrameric or hexameric oligomer structure formed by association of common dimeric components. We determined the crystal structure of E134A mutant NDK from Halomonas sp. 593 (HaNDK) and found that two kinds of tetrameric assemblies, Type I seen in the Myxococcus NDK tetramer and Type II seen in the E.coli NDK tetramer, appeared in the asymmetric unit. Change in the assembly form may be an effective way for NDK to acquire molecular characteristics suited to various circumstances.
 -Lactamase derived from
-Lactamase derived from  sp.560
sp.560Arai, Shigeki; Yonezawa, Yasushi; Okazaki, Nobuo; Tamada, Taro; Tokunaga, Hiroko*; Ishibashi, Matsujiro*; Tokunaga, Masao*; Kuroki, Ryota
no journal, ,
We determined the crystal structure of halophilic  -Lactamase obtained from
-Lactamase obtained from  sp.560 by X-ray crystallography. Since halophilic enzymes can bind many metal ions on its molecular surface, we are tring to find the Na
sp.560 by X-ray crystallography. Since halophilic enzymes can bind many metal ions on its molecular surface, we are tring to find the Na , Mg
, Mg and Cs
and Cs binding sites of halophilic
binding sites of halophilic  -Lactamase from determined crystal structure.
-Lactamase from determined crystal structure.
 -Lactamase derived from
-Lactamase derived from  sp.560
sp.560Arai, Shigeki; Tokunaga, Hiroko*; Tamada, Taro; Yonezawa, Yasushi; Adachi, Motoyasu; Yamada, Mitsugu; Ishibashi, Matsujiro*; Tokunaga, Masao*; Kuroki, Ryota
no journal, ,
Halophilic proteins can bind various inorganic ions with its negative charges located over the molecular surface. We are investigating the molecular structure of the halophilic proteins because halophilic proteins might be used as a material capturing for the rare metals and/or the radioactive metal ions. Recently, we succeeded in X-ray crystallographic analysis of the halophilic  -Lactamase (HaBLA) derived from
-Lactamase (HaBLA) derived from  sp.560 at 3.0
sp.560 at 3.0  resolution. From this structural analysis, we found that the back bone structure and the positively charged active site feature of HaBLA was similar to those of non-halophilic BLA. The molecular surface of HaBLA were, however, occupied by large number of negative charges. This structural information is very useful to improve the specificity of metals such as Cs or Sr.
resolution. From this structural analysis, we found that the back bone structure and the positively charged active site feature of HaBLA was similar to those of non-halophilic BLA. The molecular surface of HaBLA were, however, occupied by large number of negative charges. This structural information is very useful to improve the specificity of metals such as Cs or Sr.
Arai, Shigeki; Adachi, Motoyasu; Kawamoto, Masahide*; Tokunaga, Hiroko*; Ishibashi, Matsujiro*; Tokunaga, Masao*; Kuroki, Ryota
no journal, ,
We attempted to discover the Cs and Sr
and Sr binding sites on the halophilic
binding sites on the halophilic  -Lactamase (HaBLA) derived from
-Lactamase (HaBLA) derived from  sp.560 by the anomalous X-ray diffraction analysis. One Cs
sp.560 by the anomalous X-ray diffraction analysis. One Cs ions in the HaBLA crystal were identified by BL7 at SAGA-LS. Three Sr
ions in the HaBLA crystal were identified by BL7 at SAGA-LS. Three Sr ions in the HaBLA crystal were identified by BL38B1 at SPring-8 and NW12A at Photon Factory. Discovered Cs
ions in the HaBLA crystal were identified by BL38B1 at SPring-8 and NW12A at Photon Factory. Discovered Cs binding site can bind Cs
binding site can bind Cs selectively from a solution containing Na
selectively from a solution containing Na /Cs
/Cs = 90 mM/10 mM. This Cs
= 90 mM/10 mM. This Cs binding site is formed by two main-chain O atoms and an aromatic ring of a side chain of Trp. An aromatic ring of Trp interacts with Cs
binding site is formed by two main-chain O atoms and an aromatic ring of a side chain of Trp. An aromatic ring of Trp interacts with Cs by the cation-
by the cation-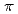 interaction. The observation of metal binding sites with relatively higher Cs
interaction. The observation of metal binding sites with relatively higher Cs selectivity provides important information that is useful for designing artificial Cs
selectivity provides important information that is useful for designing artificial Cs binding sites.
binding sites.
Arai, Shigeki; Yonezawa, Yasushi*; Adachi, Motoyasu; Tamada, Taro; Tokunaga, Hiroko*; Ishibashi, Matsujiro*; Tokunaga, Masao*; Kuroki, Ryota
no journal, ,
Halophilic proteins have unique structural characteristics: high content of acidic residues creating negatively charged surface, high reversibility of tertiary structure and activity even in high salt concentration, which make it possible to create potent adhesives for metal ions. As part of our structure-function studies for halophilic proteins, we succeeded in crystallization of several halophilic proteins using divalent metal ions as additives. Here, we show successful examples of structural determination of three halophilic proteins: two are alkaline phosphatase from  sp.593 (HaAP) and
sp.593 (HaAP) and  -Lactamase from
-Lactamase from  sp.560 (HaBLA) determined to 2.1
sp.560 (HaBLA) determined to 2.1 and 2.0
and 2.0 resolution, respectively, and the other is nucleoside diphosphate kinase from
resolution, respectively, and the other is nucleoside diphosphate kinase from  sp.593 (HaNDK) determined to 2.3
sp.593 (HaNDK) determined to 2.3 resolution.
resolution.
 -lactamase from moderate halophile
-lactamase from moderate halophileArai, Shigeki; Adachi, Motoyasu; Tamada, Taro; Tokunaga, Hiroko*; Ishibashi, Matsujiro*; Tokunaga, Masao*; Kuroki, Ryota
no journal, ,
A high content of acidic residues of halophilic proteins may destabilize the structure of halophilic protein and take away an enzymatic function under low salt concentration. However, periplasmic proteins of moderate halophiles require adapting a wide range of salt concentration (0.5 M - saturated NaCl). In this study, we investigated the structural and functional characteristics of HaBLA derived from periplasm of a moderate halophile  sp.560. By isothermal titration calorimetry, it was clarified that HaBLA hydrolyses penicillin G under 0 - 4 M NaCl. Moreover, by X-ray crystallographic analysis, we found the positive charges and the hydrophobic cluster near the entrance of the active site of HaBLA. These structural characteristics may attract penicillin G to the active site of HaBLA, which may participate in the low salt concentration adaptation of HaBLA.
sp.560. By isothermal titration calorimetry, it was clarified that HaBLA hydrolyses penicillin G under 0 - 4 M NaCl. Moreover, by X-ray crystallographic analysis, we found the positive charges and the hydrophobic cluster near the entrance of the active site of HaBLA. These structural characteristics may attract penicillin G to the active site of HaBLA, which may participate in the low salt concentration adaptation of HaBLA.
 sp.593 (HaNDK)
sp.593 (HaNDK)Arai, Shigeki; Yonezawa, Yasushi; Okazaki, Nobuo; Tamada, Taro; Tokunaga, Hiroko*; Ishibashi, Matsujiro*; Tokunaga, Masao*; Kuroki, Ryota
no journal, ,
The nucleoside diphosphate kinases (NDKs) are known to have a tetrameric or hexameric oligomer structure formed by association of common dimeric components. In order to understand the oligomeric structure of NDK, the wild-type NDK from Halomonas sp. 593 (HaNDK) was determined by X-ray crystallography. The wild-type HaNDK only exhibited a dimeric form that is the same as a common dimer unit seen in other NDKs. This is the first evidence of a dimeric structure for HaNDK. To explore the effect of mutation on the assembly form of HaNDK, E134, which is a key interface residue for other tetrameric NDKs, was replaced with Ala, and the structure was determined by X-ray crystallography. Two kinds of tetrameric assemblies, Type I and Type II, appeared in the asymmetric unit of the E134A mutant HaNDK. The change from dimeric to tetrameric assembly is attributed to the removal of negative charge repulsion caused by the E134 in the wild-type HaNDK.
Arai, Shigeki; Yonezawa, Yasushi*; Tamada, Taro; Tokunaga, Hiroko*; Ishibashi, Matsujiro*; Tokunaga, Masao*; Kuroki, Ryota
no journal, ,
no abstracts in English
 and Sr
and Sr recognition mechanism of halophilic
recognition mechanism of halophilic  -lactamase
-lactamaseArai, Shigeki; Yonezawa, Yasushi; Adachi, Motoyasu; Tokunaga, Hiroko*; Ishibashi, Matsujiro*; Tokunaga, Masao*; Kuroki, Ryota
no journal, ,
Proteins can distinguish various metal ions. Recently, we succeeded in X-ray crystallographic analysis of the halophilic  -Lactamase (HaBLA) derived from
-Lactamase (HaBLA) derived from  sp.560 under the existence of Cs+ and Sr2+. Diffraction data at 2.0
sp.560 under the existence of Cs+ and Sr2+. Diffraction data at 2.0  resolution (space group P31, Unit cell a = b =115.9
resolution (space group P31, Unit cell a = b =115.9  , c =67.9
, c =67.9  , Rmerge 9.6%) was collected. Three Cs
, Rmerge 9.6%) was collected. Three Cs and six Sr
and six Sr metal ion binding sites of HaBLA molecules in the asymmetric unit were identified. This structural information is very useful to create the artificial Cs
metal ion binding sites of HaBLA molecules in the asymmetric unit were identified. This structural information is very useful to create the artificial Cs or Sr
or Sr binding site on the protein molecules.
binding site on the protein molecules.