Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Nakagawa, Hiroshi; Jochi, Yasumasa*; Kitao, Akio*; Yamamuro, Osamu*; Kataoka, Mikio*
Biophysical Journal, 117(2), p.229 - 238, 2019/07
Times Cited Count:7 Percentile:24.38(Biophysics)Softness and rigidity of proteins are reflected in the structural dynamics, which are in turn affected by the environment. The characteristic low-frequency vibrational spectrum of a protein, known as boson peak, is an indication of the structural rigidity of the protein at cryogenic temperature or dehydrated conditions. In this paper, the effect of hydration, temperature, and pressure on the boson peak and volumetric properties of a globular protein are evaluated by using inelastic neutron scattering and molecular dynamics simulation. Hydration, pressurization, and cooling shift the boson peak position to higher energy and depress the peak intensity and decreases the protein and cavity volumes, although pressure hardly affects the boson peak of the fully hydrated protein. A decrease of each volume means the increase of rigidity, which is the origin of the boson peak shift. The boson peak profile can be predicted by the total cavity volume. This prediction is effective for the evaluation of the net quasielastic scattering of incoherent neutron scattering spectra when the boson peak cannot be distinguished experimentally because of a strong contribution from quasielastic scattering.
Tokuhisa, Atsushi*; Arai, Junya*; Jochi, Yasumasa*; Ono, Yoshiyuki*; Kameyama, Toyohisa*; Yamamoto, Keiji*; Hatanaka, Masayuki*; Gerofi, B.*; Shimada, Akio*; Kurokawa, Motoyoshi*; et al.
Journal of Synchrotron Radiation, 20(6), p.899 - 904, 2013/11
Times Cited Count:5 Percentile:27.82(Instruments & Instrumentation)Nakagawa, Hiroshi; Jochi, Yasumasa*; Kitao, Akio*; Kataoka, Mikio
Biophysical Journal, 95(6), p.2916 - 2923, 2008/09
Times Cited Count:51 Percentile:78.12(Biophysics)Jochi, Yasumasa*; Nakagawa, Hiroshi; Kataoka, Mikio; Kitao, Akio*
Biophysical Journal, 94(11), p.4435 - 4443, 2008/06
Times Cited Count:24 Percentile:52.05(Biophysics)Hydration effects on protein dynamics were investigated by comparing the frequency dependence of the calculated neutron scattering spectra between full and minimal hydration states at temperatures between 100 and 300 K. The protein boson peak is observed in the frequency range 1-4 meV at 100 K in both states. The peak frequency in the minimal hydration state shifts to lower than that in the full hydration state. Protein motions with frequency higher than 4 meV were shown to undergo almost harmonic motion in both states at all temperatures simulated, whereas those with frequency lower than 1 meV dominate the total fluctuations above 220 K and contribute to the origin of the glass-like transition. At 300 K, the boson peak becomes buried in the quasi-elastic contributions in the full hydration state, but is still observed in the minimal hydration state. The boson peak is observed when protein dynamics are trapped within a local minimum of its energy surface. Protein motions, which contribute to the boson peak, are distributed throughout the whole protein. Fine structure of the dynamics structure factor is expected to be detected by the experiment if a high resolution instrument is developed in the near future.
Jochi, Yasumasa*; Nakagawa, Hiroshi; Kataoka, Mikio; Kitao, Akio*
Journal of Physical Chemistry B, 112(11), p.3522 - 3528, 2008/03
Times Cited Count:11 Percentile:26.89(Chemistry, Physical)Molecular dynamics simulations of crystalline Staphylococcal nuclease in full and minimal hydration states were performed to study hydration effects on protein dynamics at temperatures ranging from 100 to 300 K. In a full hydration state (hydration ratio in weight, h = 0.49), gaps are fully filled with water molecules, whereas only crystal waters are included in a minimal hydration state (h = 0.09). The inflection of the atomic mean-square fluctuation of protein as a function of temperature, known as the glass-like transition, is observed at 220 K in both cases, which is more significant in the full hydration state. By examining the temperature dependence of residual fluctuation, we found that the increase of fluctuations in the loop and terminal regions, which are exposed to water, is much greater than in other regions in the full hydration state, but the mobility of the corresponding regions are relatively restricted in the minimal hydration state by inter-molecular contact. The atomic mean-square fluctuation of water molecules in the full hydration state at 300 K is one order of magnitude greater than that in the minimal hydration state. Above the transition temperature, most water molecules in the full hydration state behave like bulk water, and act as a lubricant for protein dynamics. In contrast, water molecules in the minimal hydration state tend to form more hydrogen bonds with the protein, restricting the fluctuation of these water molecules to the level of the protein. Thus, inter-molecular interaction and solvent mobility are important to understand the glass-like transition in proteins.
Tokuhisa, Atsushi; Jochi, Yasumasa*; Nakagawa, Hiroshi; Kitao, Akio*; Kataoka, Mikio
Physical Review E, 75(4), p.041912_1 - 041912_8, 2007/05
Times Cited Count:21 Percentile:67.23(Physics, Fluids & Plasmas)Elastic incoherent neutron scattering (EINS) data can be approximated with a Gaussian function of q in a low q region. However, in a higher q region the deviation from a Gaussian function becomes non-negligible. Protein dynamic properties can be derived from the analyses of the non-Gaussian behavior, which has been experimentally investigated. To evaluate the origins of the non-Gaussian behavior of protein dynamics, we conducted a molecular dynamics (MD) simulation of Staphylococcal nuclease. Instead of the ordinary cumulant expansion, we decomposed the non-Gaussian terms into three components: (1) the component originating from the heterogeneity of the mean-square fluctuation, (2) that from the anisotropy, and (3) that from higher order terms such as anharmonicity. The MD simulation revealed various dynamics for each atom. The atomic motions are classified into three types: (1) "harmonic", (2) "anisotropic", and (3) "anharmonic". However, each atom has a different degree of anisotropy. The contribution of the anisotropy to the total scattering function averages out due to these differences. Anharmonic motion is described as the jump among multiple minima. The jump distance and the probability of the residence at one site vary from atom to atom. Each anharmonic component oscillates between positive and negative values. Thus, the contribution of the anharmonicity to the total scattering is canceled due to the variations in the anharmonicity. Consequently, the non-Gaussian behavior of the total EINS from a protein can be analyzed by the dynamical heterogeneity.
Ishida, Hisashi; Higuchi, Mariko; Yonetani, Yoshiteru*; Kano, Takuma; Jochi, Yasumasa*; Kitao, Akio*; Go, Nobuhiro
Annual Report of the Earth Simulator Center April 2005 - March 2006, p.237 - 240, 2007/01
no abstracts in English
Nakagawa, Hiroshi; Tokuhisa, Atsushi*; Kamikubo, Hironari*; Jochi, Yasumasa*; Kitao, Akio*; Kataoka, Mikio*
Materials Science & Engineering A, 442(1-2), p.356 - 360, 2006/12
Times Cited Count:4 Percentile:33.36(Nanoscience & Nanotechnology)The dynamical heterogeneity of a globular soluble protein was studied by elastic incoherent neutron scattering and molecular simulations. The q-dependence of the elastic incoherent neutron scattering shows a non-Gaussianity, a deviation from Gaussian approximation. We determined that the dynamical heterogeneity explains the non-Gaussianity, although the anharmonicity is also plausible origin. Molecular dynamics simulations confirmed that the non-Gaussianity is mainly due to the dynamical heterogeneity at a lower energy resolution, 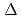
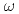 =1meV. On the other hand, the contribution from the anharmonicities to the non-Gaussianity became substantial at a higher resolution,
=1meV. On the other hand, the contribution from the anharmonicities to the non-Gaussianity became substantial at a higher resolution, 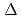
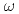 =10
=10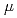 eV. Regardless, the dynamical heterogeneity is the dominant factor for the non-Gaussianity.
eV. Regardless, the dynamical heterogeneity is the dominant factor for the non-Gaussianity.
Nakagawa, Hiroshi; Kataoka, Mikio*; Jochi, Yasumasa*; Kitao, Akio*; Shibata, Kaoru; Tokuhisa, Atsushi*; Tsukushi, Itaru*; Go, Nobuhiro
Physica B; Condensed Matter, 385-386(2), p.871 - 873, 2006/11
Times Cited Count:14 Percentile:53.07(Physics, Condensed Matter)The boson peak of a protein was examined in relation to hydration using staphylococcal nuclease. Although the boson peak is commonly observed in synthetic polymers, glassy materials and amorphous materials, the origin of the boson peak is not fully understood. The motions that contribute to the peak are harmonic vibrations. Upon hydration the peak frequency shifts to a higher frequency and the effective force constant of the vibration increases at low temperatures, suggesting that the protein energy surface is modified. Hydration of the protein leads to a more rugged surface and the vibrational motions are trapped within the local minimum at cryogenic temperatures. The origin of the protein boson peak may be related to this rugged energy surface.
Ishida, Hisashi; Higuchi, Mariko; Yonetani, Yoshiteru*; Kano, Takuma; Jochi, Yasumasa*; Kitao, Akio*; Go, Nobuhiro
Annual Report of the Earth Simulator Center April 2004 - March 2005, p.241 - 246, 2005/12
no abstracts in English
Jochi, Yasumasa*; Kitao, Akio*; Go, Nobuhiro
Journal of the American Chemical Society, 127(24), p.8705 - 8709, 2005/06
Times Cited Count:27 Percentile:60.62(Chemistry, Multidisciplinary)no abstracts in English
Ishida, Hisashi; Jochi, Yasumasa*; Higuchi, Mariko; Kano, Takuma; Kitao, Akio*; Go, Nobuhiro
Annual Report of the Earth Simulator Center April 2003 - March 2004, p.175 - 179, 2004/07
no abstracts in English
Nakagawa, Hiroshi; Kataoka, Mikio; Jochi, Yasumasa*; Kitao, Akio*; Yamamuro, Osamu*
no journal, ,
no abstracts in English
Nakagawa, Hiroshi; Jochi, Yasumasa*; Kitao, Akio*; Kataoka, Mikio
no journal, ,
To understand the effect of hydration on protein dynamics, inelastic neutron scattering experiments were carried out on Staphylococcal nuclease samples at differing hydration levels: dehydrated, partially hydrated and hydrated. At cryogenic temperatures, hydration affected the collective motions with energies lower than 5 meV, while the high energy localized motions were independent of hydration. The prominent change was a shift of boson peak toward higher energy by hydration, suggesting hardening of harmonic potential at local minima on the energy landscape. The 240 K transition was observed only for the hydrated protein. Significant quasi-elastic scattering at 300 K was observed only for the hydrated sample, indicating that the origin of the transition is the motion activated by hydration water. The neutron scattering profile of the partially hydrated sample was quite similar to that of the hydrated sample at 100 and 200 K, while it was close to the dehydrated sample at 300 K, indicating that partial hydration is sufficient to affect the harmonic nature of protein dynamics, and that there is a threshold hydration level to activate the anharmonic motions. Thus, hydration water controls both the harmonic and anharmonic protein dynamics, by differing means.
Nakagawa, Hiroshi; Kataoka, Mikio; Jochi, Yasumasa*; Yamamuro, Osamu*; Nakajima, Kenji; Kawamura, Seiko
no journal, ,
Proteins are functional elements in all living organisms. Almost all vital phenomena are mediated by specific proteins. A protein is a hetero-polymer comprised of 20 types of amino acids. The sequence of amino acids of a protein is strictly determined by genetic code. When the polypeptide chain with the amino acid sequence encoded in the gene is biosynthesized, the polypeptide chain folds spontaneously into the unique tertiary structure. The tertiary structure of a protein can be determined by X-ray crystallography. Protein works in an aqueous environment at ambient temperature, indicating that protein cannot escape from thermal fluctuations. In fact, proteins are fluctuating thermally and can take some conformational substates. The magnitude of physiologically relevant input is as the same level as the thermal fluctuations. The understanding of protein dynamics is important to clarify how protein can discriminate physiologically relevant motions from random fluctuation. It is generally recognized that the internal motions of protein is essential for a protein function. Protein internal dynamics is characterized in the time scale of pico - nano second and in the space scale of the order of angstrom. Inelastic neutron scattering (INS) measurement is a powerful and unique technique for studying the protein dynamics. Here, we showed the protein dynamics studies using AGNES and AMATERAS spectrometer, which are installed at JRR-3 and J-PARC, respectively. We will discuss the hydration, temperature and pressure effect on protein dynamics as well as the difference of dynamics between folded and unfolded protein.
Nakagawa, Hiroshi; Jochi, Yasumasa*; Yamamuro, Osamu*; Kataoka, Mikio
no journal, ,
Protein dynamics is closely related with protein biological function. Protein dynamical transition and boson peak is observed by neutron inelastic scattering and molecular dynamics simulation. Protein volume fluctuation is related with cavity in protein structure. The cavity volume is affected by pressure. It is important to examine the effect of pressure on protein dynamics. We examined the effect of pressure on protein dynamical transition and boson peak of Staphylococcal nuclease by neutron inelastic scattering at AGNES spectrometer at JRR-3. We found that boson peak shifted to higher energy upon pressure at 900 MP. And quasi-elastic scattering is restricted by pressure.
Nakagawa, Hiroshi; Jochi, Yasumasa*; Yamamuro, Osamu*; Kataoka, Mikio
no journal, ,
Intrinsically disordered protein (IDP) folds from disordered structure into folded structure upon binding of ligand. In this folding reaction, the structural fluctuation plays an important role for the recognition of target molecules. In the physiological solution, protein stability and dynamics is affected by hydration. It is essential to understand the hydration and dynamics of IDP for elucidation of biological function of IDP. Fragment mutant of Staphylococcal nuclease (SNase) is denatured state at physiological condition and fold upon binding of ligand, therefore, is good model for IDP. We found the difference of dynamics and hydration between folded and unfolded state of SNase by using inelastic neutron scattering experiment.
Nakagawa, Hiroshi; Jochi, Yasumasa*; Kitao, Akio*; Kataoka, Mikio
no journal, ,
Nakagawa, Hiroshi; Jochi, Yasumasa*; Kitao, Akio*; Kataoka, Mikio
no journal, ,
Nakagawa, Hiroshi; Jochi, Yasumasa*; Kitao, Akio*; Kataoka, Mikio
no journal, ,
no abstracts in English