Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Nakagawa, Hiroshi; Kamikubo, Hironari*; Kataoka, Mikio
Biochimica et Biophysica Acta; Proteins and Proteomics, 1804(1), p.27 - 33, 2010/01
Times Cited Count:19 Percentile:45.30(Biochemistry & Molecular Biology)In order to examine the properties specific to the folded protein, the effect of the conformational states on protein dynamical transition was studied by incoherent elastic neutron scattering for both wild type and a deletion mutant of staphylococcal nuclease. The deletion mutant of SNase which lacks C-terminal 13 residues takes a compact denatured structure, and can regard as a model of intrinsic unstructured protein. Incoherent elastic neutron scattering experiments were carried out at various temperature between 10K and 300K on IN10 and IN13 installed at ILL. Temperature dependence of mean square displacements was obtained by the q-dependence of elastic scattering intensity. The measurements were performed on dried and hydrated powder samples. No significant differences were observed between wild type and the mutant for the hydrated samples, while significant differences were observed for the dried samples. A dynamical transition at 140K observed for both dried and hydrated samples. The slopes of the temperature dependence of MSD before transition and after transition are different between wild type and the mutant, indicating the folding induces hardening. The hydration water activates a further transition at 240K. The behavior of the temperature dependence of MSD is indistinguishable for wild type and the mutant, indicating that hydration water dynamics dominate the dynamical properties.
Yamaguchi, Shigeo*; Kamikubo, Hironari*; Kurihara, Kazuo; Kuroki, Ryota; Niimura, Nobuo*; Shimizu, Nobutaka*; Yamazaki, Yoichi*; Kataoka, Mikio*
Proceedings of the National Academy of Sciences of the United States of America, 106(2), p.440 - 444, 2009/01
Times Cited Count:163 Percentile:94.50(Multidisciplinary Sciences)Yamaguchi, Shigeo*; Kamikubo, Hironari*; Shimizu, Nobutaka*; Yamazaki, Yoichi*; Imamoto, Yasushi*; Kataoka, Mikio
Photochemistry and Photobiology, 83(2), p.336 - 338, 2007/03
Times Cited Count:7 Percentile:16.57(Biochemistry & Molecular Biology)The exact positions of all the hydrogen atoms in photoactive yellow protein (PYP) is important for understanding the molecular mechanism of the photoreaction because the protonation/deprotonation of certain amino acid residues and rearrangements in the hydrogen bond network are involved in the conformational changes of PYP. Neutron crystallography is one of the most effective methods to determine the hydrogen positions. However, a large crystal is required for neutron crystallography because a neutron incident flux is quite limited. In addition, the crystal should be grown from heavy water to reduce the incoherent background from hydrogen. We prepared a large crystal of PYP (dimensions: 1.5  0.7
0.7  0.7 mm
0.7 mm ) for neutron crystallography using ammonium sulfate with sodium chloride. The obtained large crystal gave X-ray diffraction spots up to 0.84
) for neutron crystallography using ammonium sulfate with sodium chloride. The obtained large crystal gave X-ray diffraction spots up to 0.84  . Although some of the hydrogen atoms could be observed in the high resolution X-ray crystal structure, functionally important hydrogen atoms were impossible to see, indicating the importance of neutron crystallography. Thus, we optimized the crystallization conditions with heavy water and successfully obtained neutron diffraction spots up to 2.1
. Although some of the hydrogen atoms could be observed in the high resolution X-ray crystal structure, functionally important hydrogen atoms were impossible to see, indicating the importance of neutron crystallography. Thus, we optimized the crystallization conditions with heavy water and successfully obtained neutron diffraction spots up to 2.1  with the crystal in D
with the crystal in D O.
O.
Nakagawa, Hiroshi; Tokuhisa, Atsushi*; Kamikubo, Hironari*; Jochi, Yasumasa*; Kitao, Akio*; Kataoka, Mikio*
Materials Science & Engineering A, 442(1-2), p.356 - 360, 2006/12
Times Cited Count:4 Percentile:33.36(Nanoscience & Nanotechnology)The dynamical heterogeneity of a globular soluble protein was studied by elastic incoherent neutron scattering and molecular simulations. The q-dependence of the elastic incoherent neutron scattering shows a non-Gaussianity, a deviation from Gaussian approximation. We determined that the dynamical heterogeneity explains the non-Gaussianity, although the anharmonicity is also plausible origin. Molecular dynamics simulations confirmed that the non-Gaussianity is mainly due to the dynamical heterogeneity at a lower energy resolution, 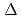
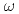 =1meV. On the other hand, the contribution from the anharmonicities to the non-Gaussianity became substantial at a higher resolution,
=1meV. On the other hand, the contribution from the anharmonicities to the non-Gaussianity became substantial at a higher resolution, 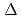
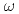 =10
=10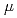 eV. Regardless, the dynamical heterogeneity is the dominant factor for the non-Gaussianity.
eV. Regardless, the dynamical heterogeneity is the dominant factor for the non-Gaussianity.
Endo, Hitoshi; Matsumoto, Atsushi; Kamikubo, Hironari*; Kataoka, Mikio
no journal, ,
Proteins are fundamental for life activities. Their functions highly depend on the structures. However, recent researches suggest that the dynamics of proteins is also important to express the functions. In this study, the structure and dynamics of Staphylococcal nuclease (SNase) in water are investigated by small-angle X-ray scattering and dynamic light scattering. SNase is one of the ideal models to study the dynamics of proteins due to the large fluctuation.
Endo, Hitoshi; Tominaga, Taiki; Takata, Shinichi; Matsumoto, Atsushi; Iwase, Hiroki*; Kamikubo, Hironari*; Kataoka, Mikio
no journal, ,
The dynamic and static structure factors for Staphylococcal Nuclease (SNase), which is a nucleolytic enzyme derived from Staphylococcus aureus, were evaluated by neutron spin echo (NSE)and small-angle neutron scattering (SANS) experiments. The SANS experiment was performed with TAIKAN (BL15) time-of-flight diffractometer at J-PARC/MLF, and we could obtain the static structure factors with wide Q range (0.2 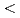 Q[1/
Q[1/![$AA] $<$$](/search/images/LRAUCXJAEQ4CI.gif) 2). The NSE measurement was performed with IN15 spectrometer at ILL. Grenoble, which enabled us to obtain intermediate scattering functions over 200 nanoseconds. The effects of hydration and internal motions were considered.
2). The NSE measurement was performed with IN15 spectrometer at ILL. Grenoble, which enabled us to obtain intermediate scattering functions over 200 nanoseconds. The effects of hydration and internal motions were considered.
Yamaguchi, Shigeo*; Kamikubo, Hironari*; Kurihara, Kazuo; Kuroki, Ryota; Niimura, Nobuo*; Shimizu, Nobutaka*; Yamazaki, Yoichi*; Kataoka, Mikio*
no journal, ,
Yamaguchi, Shigeo*; Kamikubo, Hironari*; Kurihara, Kazuo; Shimizu, Tetsuya*; Yamazaki, Yoichi*; Kuroki, Ryota; Niimura, Nobuo*; Kataoka, Mikio*
no journal, ,