Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Nakagawa, Hiroshi; Yonetani, Yoshiteru*; Nakajima, Kenji; Kawamura, Seiko; Kikuchi, Tatsuya*; Inamura, Yasuhiro; Kataoka, Mikio*; Kono, Hidetoshi*
JPS Conference Proceedings (Internet), 33, p.011101_1 - 011101_6, 2021/03
Hydration water dynamics were measured by quasi-elastic neutron scattering with Hn O/D
O/D O contrast for two DNA dodecamers, 5'CGCG
O contrast for two DNA dodecamers, 5'CGCG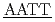 CGCG'3 and 5'CGCG
CGCG'3 and 5'CGCG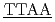 CGCG'3, which have been computationally shown to be structurally rigid and flexible, respectively. The dynamical transitions of the hydration water as well as DNA were observed for both sequences at approximately 240 K. Above the transition temperature, the mean square displacements of the hydration water for the rigid sequence were smaller than those for the flexible one. Furthermore, the relaxation time of the hydration water was longer in the rigid DNA than in the flexible DNA. We suggest that hydration water dynamics on the picosecond timescale are associated with sequence-dependent deformability of DNA.
CGCG'3, which have been computationally shown to be structurally rigid and flexible, respectively. The dynamical transitions of the hydration water as well as DNA were observed for both sequences at approximately 240 K. Above the transition temperature, the mean square displacements of the hydration water for the rigid sequence were smaller than those for the flexible one. Furthermore, the relaxation time of the hydration water was longer in the rigid DNA than in the flexible DNA. We suggest that hydration water dynamics on the picosecond timescale are associated with sequence-dependent deformability of DNA.
Nakagawa, Hiroshi; Kataoka, Mikio*
Biochimica et Biophysica Acta; General Subjects, 1864(4), p.129536_1 - 129536_6, 2020/04
Times Cited Count:3 Percentile:18.03(Biochemistry & Molecular Biology)The rigidity and flexibility of a protein is reflected in its structural dynamics. Studies on protein dynamics often focus on flexibility and softness; this review focuses on protein structural rigidity. The extent of rigidity can be assessed experimentally with incoherent neutron scattering; a method that is complementary to molecular dynamics simulation. This experimental technique can provide information about protein dynamics in timescales of pico- to nanoseconds and at spatial scales of nanometers; these dynamics can help quantify the rigidity of a protein by indices such as force constant, Boson peak, dynamical transition, and dynamical heterogeneity. These indicators also reflect the rigidity of a protein's secondary and tertiary structures. In addition, the indices reveal how rigidity is influenced by different environmental parameters, such as hydration, temperature, pressure, and protein-protein interactions. Hydration affects both rigidity and softness more than other environmental factors. Interestingly, hydration affects harmonic and anharmonic motions in opposite ways. This difference is probably due to the protein's dynamic coupling with water molecules via hydrogen bonding.
Nakagawa, Hiroshi; Jochi, Yasumasa*; Kitao, Akio*; Yamamuro, Osamu*; Kataoka, Mikio*
Biophysical Journal, 117(2), p.229 - 238, 2019/07
Times Cited Count:3 Percentile:13.03(Biophysics)Softness and rigidity of proteins are reflected in the structural dynamics, which are in turn affected by the environment. The characteristic low-frequency vibrational spectrum of a protein, known as boson peak, is an indication of the structural rigidity of the protein at cryogenic temperature or dehydrated conditions. In this paper, the effect of hydration, temperature, and pressure on the boson peak and volumetric properties of a globular protein are evaluated by using inelastic neutron scattering and molecular dynamics simulation. Hydration, pressurization, and cooling shift the boson peak position to higher energy and depress the peak intensity and decreases the protein and cavity volumes, although pressure hardly affects the boson peak of the fully hydrated protein. A decrease of each volume means the increase of rigidity, which is the origin of the boson peak shift. The boson peak profile can be predicted by the total cavity volume. This prediction is effective for the evaluation of the net quasielastic scattering of incoherent neutron scattering spectra when the boson peak cannot be distinguished experimentally because of a strong contribution from quasielastic scattering.
Nakagawa, Hiroshi; Kataoka, Mikio*
Biophysics and Physicobiology (Internet), 16, p.213 - 219, 2019/00
Incoherent neutron scattering (INS) is one of the useful experimental methods for studying protein dynamics at the pico-nanosecond timescale. At this timescale, protein dynamics is highly coupled with hydration, which is observed as protein dynamical transition (PDT). INS is very sensitive to hydrogen atomic dynamics because of the large incoherent scattering cross section of hydrogen atom, and thus, the INS of a hydrated protein provides overall dynamic information about the protein, including hydration water. Separation of hydration water dynamics is essential for understanding hydration-related protein dynamics. H O/D
O/D O exchange is an effective method in the context of INS experiments for observing the dynamics of protein and hydration water separately. Neutron scattering is directly related to the van Hove space-time correlation function, which can be calculated quantitatively by performing molecular dynamics (MD) simulations. Diffusion and hydrogen bond dynamics of hydration water can be analyzed by performing MD simulation. MD simulation is useful for analyzing the dynamic coupling mechanism in hydration-related protein dynamics from the viewpoint of interpreting INS data because PDT is induced by hydration. In the present work, we demonstrate the methodological advantages of the H
O exchange is an effective method in the context of INS experiments for observing the dynamics of protein and hydration water separately. Neutron scattering is directly related to the van Hove space-time correlation function, which can be calculated quantitatively by performing molecular dynamics (MD) simulations. Diffusion and hydrogen bond dynamics of hydration water can be analyzed by performing MD simulation. MD simulation is useful for analyzing the dynamic coupling mechanism in hydration-related protein dynamics from the viewpoint of interpreting INS data because PDT is induced by hydration. In the present work, we demonstrate the methodological advantages of the H O/D
O/D O exchange technique in INS and the compatibility of INS and MD simulation as tools for studying protein dynamics and hydration water.
O exchange technique in INS and the compatibility of INS and MD simulation as tools for studying protein dynamics and hydration water.
Nakagawa, Hiroshi; Kataoka, Mikio*
Shiki, 36, P. 6, 2017/09
We report that the degree of bendability of DNA molecules with double helical structure depends on the base sequence and demonstrated by prediction by computer and so on by demonstrating by neutron quasielastic scattering.
Nakagawa, Hiroshi; Kataoka, Mikio*
Kasokuki, 13(4), p.214 - 219, 2017/01
Life science is expected to be one of the major subjects of neutron research. The quantum beam properties of neutron, isotope effect and inelastic/quasi-elastic scattering are useful for studying crystal structures, solution structures and dynamic structures of biomolecules. Research on physical properties of biological materials by inelastic scattering is promising as applied research such as food science. The high-intensity pulse neutrons realized by the accelerator not only has developed the life science research so far, but also is opening a gate to the academic field which has never been used before.
Nakagawa, Hiroshi; Kataoka, Mikio*
Radioisotopes, 64(10), p.647 - 659, 2015/10
Inelastic neutron scattering can measure the protein thermal fluctuations under the physiological aqueous environment, especially it is powerful to observe the low-energy protein dynamics in THz region, which are revealed theoretically to be coupled with solvations. Neutron makes possible the selective observation of protein and hydration water by deuteration. The complementary analysis with molecular dynamics simulation is also effective for the study of protein hydration. Some examples of the application toward the understanding of molecular basis of protein functions will be introduced.
Nakagawa, Hiroshi; Kataoka, Mikio*
Reito, 90(1054), p.569 - 573, 2015/08
Incoherent neutron scattering (INS) can measure the protein thermal fluctuations under the physiological aqueous environment. The protein dynamics should be characterized with wide time-space ranges. INS is a promising experimental method for analysis of water mobility and protein dynamical transition. Neutron has high permeability in materials, and can observe non-destructively the molecular dynamics. Neutron can give information about vibrational density of states, relaxation and diffusion processes, and mean-square displacements. Molecular Dynamics (MD) simulation gives information on the molecular structure and dynamics with atomic scale, which is complementary to INS. We revealed that the hydration level dependence of the onset of protein dynamical transition is correlated with the percolation transition of hydration water. At the hydration level above the percolation transition, the hydration water dynamics change significantly above the protein dynamical transition temperature. Protein dynamical transition is coupled with the fluctuation of the hydration water network covered over the protein surface.
Nakagawa, Hiroshi; Kataoka, Mikio*
Hamon, 24(特別号), 2 Pages, 2014/11
Neutron beam also treats the water molecule even an electron beam or X-ray can't grasp clearly. The graceful food texture of the Ibaraki specialty "drying potato" stands up by an exquisite balance of shelf stability and gusto. As well as the simple moisture content, it's said that in what kind of state a water molecule combines with a substance influences gusto and shelf stability. "Neutron inelastic scattering" is accumulating the new knowledge of gusto's and an appetite's being safe as well as an analysis's of a protein and a DNA and is having big hope.
Nakagawa, Hiroshi; Yonetani, Yoshiteru; Nakajima, Kenji; Kawamura, Seiko; Kikuchi, Tatsuya; Inamura, Yasuhiro; Kataoka, Mikio; Kono, Hidetoshi
Physical Review E, 90(2), p.022723_1 - 022723_11, 2014/08
Times Cited Count:10 Percentile:55.34(Physics, Fluids & Plasmas)Molecular dynamics (MD) simulations and Quasi-Elastic Neutron Scattering (QENS) experiments were conducted on hydrated two DNA dodecamers with distinct deformability; 5'CGCGAATTCGCG3' and 5'CGCGTTAACGCG3'. The former is known to be rigid and the latter to be flexible. The mean-square displacements (MSDs) of DNA dodecamers exhibit so-called dynamical transition around 200-240 K for both sequences. To investigate the DNA sequence dependent dynamics, the dynamics of DNA and hydration water above the transition temperature were examined using both MD simulations and QENS experiments. The fluctuation amplitude of the AATT central tetramer is smaller, and its relaxation time is longer, than that observed in TTAA, suggesting that the AT step is kinetically more stable than TA. The sequence-dependent local base pair step dynamics correlate with the kinetics of breaking the hydrogen bond between DNA and hydration water. The sequence dependent DNA base pair step fluctuations appear above the dynamical transition temperature. Together with these results, we conclude that DNA deformability is related to the local dynamics of base pair step, themselves coupled to hydration water in the minor groove.
Nakagawa, Hiroshi; Kataoka, Mikio
Nihon Shokuhin Kagaku Kogakkai-Shi, 61(7), p.323 - 328, 2014/07
Times Cited Count:0 Percentile:0.09(Food Science & Technology)Low water activity and glass state in dried food restrict the growth of microorganisms and increase the food stability. Vitrification of food is related with the texture. Thus, hydration and vitrification is an essential factor determining the physical property and quality of food. Inelastic neutron scattering (INS) is a promising experimental method for analysis of water mobility and glass transition of food materials. Neutron can give information about vibrational density of states, relaxation and diffusion processes, and mean-square displacements. Molecular Dynamics (MD) simulation gives the information on the molecular structure and dynamics with atomic scale, which is complementary to INS. It was found that the hydration level dependence of the onset of protein glass transition is correlated with the percolation transition of hydration water. The percolation of hydration water induces the protein glass transition.
Nakagawa, Hiroshi; Kataoka, Mikio
Nihon Setchaku Gakkai-Shi, 49(11), p.427 - 432, 2013/11
Inelastic neutron scattering is useful for analysis of hydration at the material interface. It can be expected that the inelastic neutron scattering is applicable to the analysis of an adhesion phenomena. In this paper, we will explain the theory of inelastic neutron scattering and show our works about the protein hydration.
Nakagawa, Hiroshi; Kataoka, Mikio
Kobunshi, 60(4), p.195 - 196, 2011/04
Protein is a biomolecule with a regular tertiary structure. The protein functions biologically by conformational change under the thermal fluctuation. Experimentally obtained neutron has a wavelength of a few angstrom and an energy of a few meV, which correspond to the distance between atoms in the molecule and an energy of the thermal fluctuation, respectively. Therefore, the neutron is useful for the determination of molecular structure by diffraction and the observation of functional protein dynamics and hydration water dynamics with pico-nano second time scale. J-PARC, new pulse neutron source at Tokai in Ibaraki, started operation. Here, we show our recent work about the dynamics of the biomolecule.
Kataoka, Mikio; Nakagawa, Hiroshi
Neutrons in Soft Matter, p.517 - 538, 2011/03
Protein works in an aqueous environment at ambient temperature, indicating that protein cannot escape from thermal fluctuations. Protein conformation is fluctuating thermally. The protein dynamics is related to the biological function. Inelastic neutron scattering is unique method that gives information about the protein dynamics in time and space axis. However, the application of neutron to biology is quite limited so far because the signal from biological samples is weak and the incident neutron flux is not necessarily sufficient. But this situation has been improving by constructing the new neutron sources, such as J-PARC and SNS. In this chapter the neutron scattering study of protein dynamics in the wide time and space range will be reviewed. The most observable phenomena such as a protein Boson peak and dynamical transition are also observed in the other soft matter materials. These studies should contribute to the soft matter physics. In the field of biophysics, these dynamical properties are important to understand the biological function of the proteins.
Kataoka, Mikio; Nakagawa, Hiroshi
Water; The Forgotten biological molecule, p.49 - 62, 2010/11
Proteins are biologically functional elements in the living organisms. Most proteins work in an aqueous environment. Here, we will review the effect of hydration on protein dynamics. We could classify the wide neutron spectrum into the hydration uncoupled and coupled modes. The former is comprised of localized motions, such as methyl rotation and CH bending, which corresponds to vibrational spectrum observed by IR region. The latter contains the collective motions spread over whole protein observed in the low energy spectral region. Significant increase of atomic mean-square displacement at around 240 K, which is dynamical transition, was induced by hydration. Hydration hardens the harmonic properties of protein while softens protein by acquisition of anharmonic motions. At the ambient temperature, hydration affects the surface shell of the protein structure, indicating the importance of the interaction between the protein surface and hydration water for the protein dynamics. The dynamical coupling of protein with hydration water contributes to the biologically functional mobility of protein.
Nakagawa, Hiroshi; Kataoka, Mikio
Journal of the Physical Society of Japan, 79(8), p.083801_1 - 083801_4, 2010/08
Times Cited Count:37 Percentile:81.77(Physics, Multidisciplinary)Protein cannot work without hydration water. In order to clarify the effect of hydration water on protein dynamics, incoherent neutron scattering and MD simulation are performed for a protein, staphylococcal nuclease. We found that the hydration level dependence of the onset of protein dynamical transition is correlated with the percolation transition of hydration water. Hydration water dynamics change above the transition as well. The percolation of hydration water is essential to activate the anharmonic motions of a protein, which are responsible for protein function.
Nakagawa, Hiroshi; Kamikubo, Hironari*; Kataoka, Mikio
Biochimica et Biophysica Acta; Proteins and Proteomics, 1804(1), p.27 - 33, 2010/01
Times Cited Count:19 Percentile:47.02(Biochemistry & Molecular Biology)In order to examine the properties specific to the folded protein, the effect of the conformational states on protein dynamical transition was studied by incoherent elastic neutron scattering for both wild type and a deletion mutant of staphylococcal nuclease. The deletion mutant of SNase which lacks C-terminal 13 residues takes a compact denatured structure, and can regard as a model of intrinsic unstructured protein. Incoherent elastic neutron scattering experiments were carried out at various temperature between 10K and 300K on IN10 and IN13 installed at ILL. Temperature dependence of mean square displacements was obtained by the q-dependence of elastic scattering intensity. The measurements were performed on dried and hydrated powder samples. No significant differences were observed between wild type and the mutant for the hydrated samples, while significant differences were observed for the dried samples. A dynamical transition at 140K observed for both dried and hydrated samples. The slopes of the temperature dependence of MSD before transition and after transition are different between wild type and the mutant, indicating the folding induces hardening. The hydration water activates a further transition at 240K. The behavior of the temperature dependence of MSD is indistinguishable for wild type and the mutant, indicating that hydration water dynamics dominate the dynamical properties.
Nakagawa, Hiroshi; Kataoka, Mikio
Hamon, 19(2), p.91 - 94, 2009/04
Effects of hydration on dynamics of Staphylococcal nuclease were examined by inelastic neutron scattering. At cryogenic temperatures, hydration affected the collective motions with energies lower than 5 meV, whereas the high-energy localized motions were independent of hydration. The frequency-upshift of the boson peak was also observed with hydration. These results suggest hardening of harmonic potential at local minima on the energy landscape. The 240 K transition was observed only for the hydrated protein, but not for the partially hydrated protein. On the other hand, partial hydration is sufficient to affect the harmonic nature of protein dynamics. The interesting and important finding is that there is a threshold hydration level to activate the anharmonic motions. Thus, hydration water controls both the harmonic and anharmonic protein dynamics by different mechanisms.
Yamaguchi, Shigeo*; Kamikubo, Hironari*; Kurihara, Kazuo; Kuroki, Ryota; Niimura, Nobuo*; Shimizu, Nobutaka*; Yamazaki, Yoichi*; Kataoka, Mikio*
Proceedings of the National Academy of Sciences of the United States of America, 106(2), p.440 - 444, 2009/01
Times Cited Count:157 Percentile:94.63(Multidisciplinary Sciences)Nakagawa, Hiroshi; Jochi, Yasumasa*; Kitao, Akio*; Kataoka, Mikio
Biophysical Journal, 95(6), p.2916 - 2923, 2008/09
Times Cited Count:49 Percentile:78.26(Biophysics)