Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
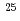 F from
F from 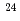 O
O ?
?Tang, T. L.*; Uesaka, Tomohiro*; Kawase, Shoichiro; Beaumel, D.*; Dozono, Masanori*; Fujii, Toshihiko*; Fukuda, Naoki*; Fukunaga, Taku*; Galindo-Uribarri, A.*; Hwang, S. H.*; et al.
Physical Review Letters, 124(21), p.212502_1 - 212502_6, 2020/05
Times Cited Count:19 Percentile:72.96(Physics, Multidisciplinary)The structure of a neutron-rich 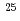 F nucleus is investigated by a quasifree (
F nucleus is investigated by a quasifree (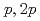 ) knockout reaction. The sum of spectroscopic factors of
) knockout reaction. The sum of spectroscopic factors of 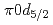 orbital is found to be 1.0
orbital is found to be 1.0 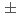 0.3. The result shows that the
0.3. The result shows that the 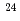 O core of
O core of 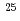 F nucleus significantly differs from a free
F nucleus significantly differs from a free 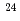 O nucleus, and the core consists of
O nucleus, and the core consists of  35%
35% 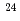 O
O , and
, and  65% excited
65% excited 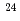 O. The result shows that the
O. The result shows that the 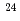 O core of
O core of 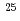 F nucleus significantly differs from a free
F nucleus significantly differs from a free 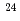 O nucleus. The result may infer that the addition of the
O nucleus. The result may infer that the addition of the 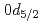 proton considerably changes the neutron structure in
proton considerably changes the neutron structure in 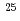 F from that in
F from that in 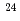 O, which could be a possible mechanism responsible for the oxygen dripline anomaly.
O, which could be a possible mechanism responsible for the oxygen dripline anomaly.
Son, N. T.*; Trinh, X. T.*; L vile, L. S.*; Svensson, B. G.*; Kawahara, Kotaro*; Suda, Jun*; Kimoto, Tsunenobu*; Umeda, Takahide*; Isoya, Junichi*; Makino, Takahiro; et al.
vile, L. S.*; Svensson, B. G.*; Kawahara, Kotaro*; Suda, Jun*; Kimoto, Tsunenobu*; Umeda, Takahide*; Isoya, Junichi*; Makino, Takahiro; et al.
Physical Review Letters, 109(18), p.187603_1 - 187603_5, 2012/11
Times Cited Count:230 Percentile:98.14(Physics, Multidisciplinary)Yamasaki, Chisato*; Murakami, Katsuhiko*; Fujii, Yasuyuki*; Sato, Yoshiharu*; Harada, Erimi*; Takeda, Junichi*; Taniya, Takayuki*; Sakate, Ryuichi*; Kikugawa, Shingo*; Shimada, Makoto*; et al.
Nucleic Acids Research, 36(Database), p.D793 - D799, 2008/01
Times Cited Count:52 Percentile:70.47(Biochemistry & Molecular Biology)Here we report the new features and improvements in our latest release of the H-Invitational Database, a comprehensive annotation resource for human genes and transcripts. H-InvDB, originally developed as an integrated database of the human transcriptome based on extensive annotation of large sets of fulllength cDNA (FLcDNA) clones, now provides annotation for 120 558 human mRNAs extracted from the International Nucleotide Sequence Databases (INSD), in addition to 54 978 human FLcDNAs, in the latest release H-InvDB. We mapped those human transcripts onto the human genome sequences (NCBI build 36.1) and determined 34 699 human gene clusters, which could define 34 057 protein-coding and 642 non-protein-coding loci; 858 transcribed loci overlapped with predicted pseudogenes.