Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
 resolution
resolutionShimizu, Noriko*; Sugiyama, Shigeru*; Maruyama, Mihoko*; Takahashi, Yoshinori*; Adachi, Motoyasu; Tamada, Taro; Hidaka, Koshi*; Hayashi, Yoshio*; Kimura, Toru*; Kiso, Yoshiaki*; et al.
Crystal Growth & Design, 10(7), p.2990 - 2994, 2010/06
Times Cited Count:11 Percentile:71.28(Chemistry, Multidisciplinary)We report crystal growth of human immunodeficiency virus 1 protease (HIV PR) in a complex with its inhibitor KNI-272 by six different methods. Comparative analysis indicates that top-seeded solution growth (TSSG) and TSSG combined with the floating and stirring technique (TSSG-FAST) are efficient strategies for rapidly obtaining large single crystals and effectively preventing polycrystallization of the seed crystal. Neutron diffraction analysis confirmed that the crystalobtained by TSSG is a high-quality single crystal. Furthermore, crystal shape was observed to be influenced by solution flow, suggesting that the degree of supersaturation significantly affects the crystal growth direction of HIV PR complex. This finding implies that the shape of the HIV PR complex crystal might be controlled by the solution flow rate.
Hidaka, Koshi*; Kimura, Toru*; Abdel-Rahman, H. M.*; Nguyen, J.-T.*; McDaniel, K. F.*; Kohlbrenner, W. E.*; Molla, A.*; Adachi, Motoyasu; Tamada, Taro; Kuroki, Ryota; et al.
Journal of Medicinal Chemistry, 52(23), p.7604 - 7617, 2009/07
Times Cited Count:20 Percentile:44.92(Chemistry, Medicinal)A series of HIV protease inhibitor based on the allophenylnorstatine structure with various P2' moieties were synthesized. Among these analogues, we discovered that a small allyl group would maintain potent enzyme inhibitory activity compared to that of the 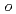 -methylbenzyl moiety in clinical candidate 1 (KNI-764, also known as JE-2147, AG-1776 or SM-319777). Introduction of an anilinic amino group to 2 (KNI-727) improved water-solubility and anti-HIV-1 activity. X-ray crystallographic analysis of 13k (KNI-1689) with a
-methylbenzyl moiety in clinical candidate 1 (KNI-764, also known as JE-2147, AG-1776 or SM-319777). Introduction of an anilinic amino group to 2 (KNI-727) improved water-solubility and anti-HIV-1 activity. X-ray crystallographic analysis of 13k (KNI-1689) with a  -methallyl group at P2' position revealed hydrophobic interactions with Ala28, Ile84, and Ile50' similar to that of 1. The presence of an additional methyl group on the allyl group in compound 13k significantly increased anti-HIV activity over 1, while providing a rational drug design for structural minimization and improving membrane permeability.
-methallyl group at P2' position revealed hydrophobic interactions with Ala28, Ile84, and Ile50' similar to that of 1. The presence of an additional methyl group on the allyl group in compound 13k significantly increased anti-HIV activity over 1, while providing a rational drug design for structural minimization and improving membrane permeability.
Adachi, Motoyasu; Ohara, Takashi; Kurihara, Kazuo; Tamada, Taro; Honjo, Eijiro; Okazaki, Nobuo; Arai, Shigeki; Shoyama, Yoshinari; Kimura, Kaname*; Matsumura, Hiroyoshi*; et al.
Proceedings of the National Academy of Sciences of the United States of America, 106(12), p.4641 - 4646, 2009/03
Times Cited Count:115 Percentile:90.49(Multidisciplinary Sciences)To further understand the catalytic mechanism and inhibitor recognition of HIV-1 protease, we need to determine the locations of key hydrogen atoms in the catalytic aspartates Asp25 and Asp125. The structure of HIV-1 protease in complex with transition-state analog KNI-272 was determined by combined neutron crystallography at 1.9  resolution and X-ray crystallography at 1.4
resolution and X-ray crystallography at 1.4  resolution. The resulting structural data shows that the catalytic residue Asp25 is protonated and that Asp125 is deprotonated. The proton on Asp25 makes a hydrogen bond with the carbonyl group of the allophenylnorstatine group in KNI-272. The deprotonated Asp125 bonds to the hydroxyl proton of Apns. The results provide direct experimental evidence for proposed aspects of the catalytic mechanism of HIV-1 protease; and can therefore contribute substantially to the development of specific inhibitors for therapeutic application.
resolution. The resulting structural data shows that the catalytic residue Asp25 is protonated and that Asp125 is deprotonated. The proton on Asp25 makes a hydrogen bond with the carbonyl group of the allophenylnorstatine group in KNI-272. The deprotonated Asp125 bonds to the hydroxyl proton of Apns. The results provide direct experimental evidence for proposed aspects of the catalytic mechanism of HIV-1 protease; and can therefore contribute substantially to the development of specific inhibitors for therapeutic application.
Matsumura, Hiroyoshi*; Adachi, Motoyasu; Sugiyama, Shigeru*; Okada, Shino*; Yamakami, Megumi*; Tamada, Taro; Hidaka, Koshi*; Hayashi, Yoshio*; Kimura, Toru*; Kiso, Yoshiaki*; et al.
Acta Crystallographica Section F, 64(11), p.1003 - 1006, 2008/11
Times Cited Count:17 Percentile:77.23(Biochemical Research Methods)This paper reports the crystallization and preliminary neutron diffraction measurements of HIV-1 protease, a potential target for anti-HIV therapy, complexed with an inhibitor (KNI-272). The aim of this neutron diffraction study is to obtain structural information about the H atoms and to determine the protonation states of the residues within the active site. The crystal was grown to a size of 1.4 mm by repeated macroseeding and a slow-cooling method using a two-liquid system. Neutron diffraction data were collected at room temperature using a BIX-4 diffractometer at the JRR-3 research reactor of the Japan Atomic Energy Agency (JAEA). The data set was integrated and scaled to 2.3
by repeated macroseeding and a slow-cooling method using a two-liquid system. Neutron diffraction data were collected at room temperature using a BIX-4 diffractometer at the JRR-3 research reactor of the Japan Atomic Energy Agency (JAEA). The data set was integrated and scaled to 2.3  resolution in space group P2(1)2(1)2, with unit-cell parameters a = 59.5, b = 87.4, c = 46.8
resolution in space group P2(1)2(1)2, with unit-cell parameters a = 59.5, b = 87.4, c = 46.8  .
.
Adachi, Motoyasu; Ohara, Takashi; Kurihara, Kazuo; Tamada, Taro; Honjo, Eijiro; Okazaki, Nobuo; Arai, Shigeki; Shoyama, Yoshinari; Matsumura, Hiroyoshi*; Sugiyama, Shigeru*; et al.
no journal, ,
We have determined a crystal structure of HIV-1 protease by neutron crystallography. The development of HIV-1 protease inhibitors is regarded as a major success of structure-based drug design and contributes to establish highly active anti-retroviral therapy for AIDS. To further understand the catalytic mechanism of HIV-1 protease and interaction between HIV-1 protease and its inhibitor, we have determined the crystal structure of HIV-1 protease in complex with a inhibitor, KNI-272 to 2.3  resolution by neutron crystallography. Our results indicates that the carbonyl group of allophenylnorstatine (Apns) in KNI-272 forms a significant hydrogen bond with protonated Asp 25, and the hydrogen atom from the hydroxyl group of Apns forms a remarkable hydrogen bond with the deprotonated Asp125. These results show direct evidence that Asp25 provides a proton to carbonyl group of substrate and Asp125 contributes to activate the attacking water molecule as a nucleophile.
resolution by neutron crystallography. Our results indicates that the carbonyl group of allophenylnorstatine (Apns) in KNI-272 forms a significant hydrogen bond with protonated Asp 25, and the hydrogen atom from the hydroxyl group of Apns forms a remarkable hydrogen bond with the deprotonated Asp125. These results show direct evidence that Asp25 provides a proton to carbonyl group of substrate and Asp125 contributes to activate the attacking water molecule as a nucleophile.
Adachi, Motoyasu; Ohara, Takashi; Kurihara, Kazuo; Tamada, Taro; Honjo, Eijiro; Okazaki, Nobuo; Arai, Shigeki; Shoyama, Yoshinari; Kimura, Kaname*; Matsumura, Hiroyoshi*; et al.
no journal, ,
HIV-1 protease is a dimeric aspartic protease that cleaves the nascent polyproteins of HIV-1 and plays an essential role in viral replication. To further understand the catalytic mechanism of HIV-1 protease, we have determined the crystal structure of HIV-1 protease in complex with a transition state mimetic tripeptide inhibitor, KNI-272 to 1.9  resolution by neutron crystallography in combination with 1.4
resolution by neutron crystallography in combination with 1.4  resolution X-ray diffraction data. Our results indicates that the carbonyl group of allophenylnorstatine in KNI-272 forms a significant hydrogen bond with protonated Asp 25, and the hydrogen atom from the hydroxyl group of Apns forms a remarkable hydrogen bond with the deprotonated Asp125. These results show direct evidence that Asp25 provides a proton to carbonyl group of substrate and Asp125 contributes to activate the attacking water molecule as a nucleophile.
resolution X-ray diffraction data. Our results indicates that the carbonyl group of allophenylnorstatine in KNI-272 forms a significant hydrogen bond with protonated Asp 25, and the hydrogen atom from the hydroxyl group of Apns forms a remarkable hydrogen bond with the deprotonated Asp125. These results show direct evidence that Asp25 provides a proton to carbonyl group of substrate and Asp125 contributes to activate the attacking water molecule as a nucleophile.
Adachi, Motoyasu; Ohara, Takashi; Kurihara, Kazuo; Tamada, Taro; Honjo, Eijiro*; Okazaki, Nobuo; Arai, Shigeki; Shoyama, Yoshinari*; Matsumura, Hiroyoshi*; Adachi, Hiroaki*; et al.
no journal, ,
To understand the catalytic mechanism of HIV-1 protease, we have determined the crystal structure of HIV-1 protease in complex with a transition state mimetic inhibitor, KNI-272 by neutron crystallography. Our results indicates that the carbonyl group of allophenylnorstatine in KNI-272 forms a significant hydrogen bond with protonated Asp 25, and the hydrogen atom from the hydroxyl group of allophenylnorstatine forms a remarkable hydrogen bond with the deprotonated Asp125. These results show direct evidence that Asp25 provides a proton to carbonyl group of substrate and Asp125 contributes to activate the attacking water molecule as a nucleophile.
Adachi, Motoyasu; Ohara, Takashi; Kurihara, Kazuo; Tamada, Taro; Honjo, Eijiro*; Okazaki, Nobuo; Arai, Shigeki; Shoyama, Yoshinari*; Matsumura, Hiroyoshi*; Sugiyama, Shigeru*; et al.
no journal, ,
In this study, we determined crystal structures of HIV-1 protease complexed with inhibitor by neutron and X-ray crystallography. Finally, we refined the structures to R-factor of 17.3% and free R-factor 20.3% by neutron crystallography and to R-factor of 10.4 % and free R-factor 12.4% by X-ray crystallography. The result shows that Asp 25 residue is protonated and Asp 125 is deprotonated. These information is important to resolve catalytic mechanism and design of new potent inhibitor.
Adachi, Motoyasu; Ohara, Takashi; Kurihara, Kazuo; Tamada, Taro; Honjo, Eijiro*; Okazaki, Nobuo; Arai, Shigeki; Shoyama, Yoshinari*; Matsumura, Hiroyoshi*; Adachi, Hiroaki*; et al.
no journal, ,
HIV-1 protease is a dimeric aspartic protease that cleaves the nascent polyproteins of HIV-1 and plays an essential role in viral replication. To further understand the catalytic mechanism of HIV-1 protease, we have determined the crystal structure of HIV-1 protease in complex with a transition state mimetic tripeptide inhibitor, KNI-272 to 1.9  ; resolution by neutron crystallography in combination with 1.4
; resolution by neutron crystallography in combination with 1.4  ; resolution X-ray diffraction data. Our results indicates that the carbonyl group of allophenylnorstatine (Apns) in KNI-272 forms a significant hydrogen bond with protonated Asp 25, and the hydrogen atom from the hydroxyl group of Apns forms a remarkable hydrogen bond with the deprotonated Asp125. These results show direct evidence that Asp25 provides a proton to carbonyl group of substrate and Asp125 contributes to activate the attacking water molecule as a nucleophile.
; resolution X-ray diffraction data. Our results indicates that the carbonyl group of allophenylnorstatine (Apns) in KNI-272 forms a significant hydrogen bond with protonated Asp 25, and the hydrogen atom from the hydroxyl group of Apns forms a remarkable hydrogen bond with the deprotonated Asp125. These results show direct evidence that Asp25 provides a proton to carbonyl group of substrate and Asp125 contributes to activate the attacking water molecule as a nucleophile.
Hidaka, Koshi*; Toda, Yuki*; Adachi, Motoyasu; Kuroki, Ryota; Kiso, Yoshiaki*
no journal, ,
Molecular dynamic simulations of the inhibitor suggested existence of additional stable bridging water molecules to support the binding with mutated proteases. To increase the numbers of bridging water molecules, we replaced amide with sulfonyl and oxamide structures at both terminals of pseudo-symmetric peptides based on HMC. In summary, oxamide modifications resulted in a moderate activity loss against lopinavir-resistant mutations compared to inhibitors without oxamide. This method to accompany multiple bridging water molecules could be applicable to design protease inhibitors to defeat the drug resistance from amino acid mutations.
Adachi, Motoyasu; Shimizu, Rumi; Kuroki, Ryota; Moriya, Keisuke*; Kidokoro, Shunichi*; Hidaka, Koshi*; Tsuda, Yuko*; Kiso, Yoshiaki*
no journal, ,
Human immune deficiency virus protease-I (HIV-PR) is one of the important drug target proteins for the acquired immune deficiency syndrome. In this study, we designed the two single chain derivatives of wild-type and A17 type HIV-PRs in which the catalytic residue of Asp25 was placed with Asn25 to inactivate the enzyme. The tertiary structure of sc-HIV-PR of wild-type and A17 type were determined by X-ray crystallography to 1.1 and 1.5  resolution, respectively. The both complex structures showed that Asn25 forms hydrogen bond with carbonyl group of inhibitor.
resolution, respectively. The both complex structures showed that Asn25 forms hydrogen bond with carbonyl group of inhibitor.
Adachi, Motoyasu; Hatanaka, Takaaki*; Ito, Yuji*; Hidaka, Koshi*; Tsuda, Yuko*; Kiso, Yoshiaki*; Kuroki, Ryota
no journal, ,
HIV protease is known as a drug target protein. It is important to clear relationship between structural data and kinetic and physicochemical parameters for drug design. We prepared single-chained enzyme linked by two amino acids and cross-liked enzyme bridged by disulfide bond. The two single-chained enzymes were expressed as inclusion body and refolded by dilution method. The purified enzyme complexed with inhibitor KNI-272 was crystallized, and solved the crystal structures. The designed sc- and cl-HIV-PR will be useful for evaluate the affinity of newly designed inhibitors from kinetic and thermodynamic point of view. Finally, we also report the results of analysis in affinity of inhibitors by surface plasmon resonance.
Adachi, Motoyasu; Tamada, Taro; Kuroki, Ryota; Hidaka, Koshi*; Kimura, Toru*; Kiso, Yoshiaki*; Yamamoto, Kohei*; Kidokoro, Shunichi*
no journal, ,
no abstracts in English
Adachi, Motoyasu; Ohara, Takashi; Kurihara, Kazuo; Tamada, Taro; Honjo, Eijiro; Okazaki, Nobuo; Arai, Shigeki; Shoyama, Yoshinari; Matsumura, Hiroyoshi*; Sugiyama, Shigeru*; et al.
no journal, ,
no abstracts in English
Adachi, Motoyasu; Arai, Shigeki; Matsumoto, Fumiko; Kuroki, Ryota; Hatanaka, Takaaki*; Ito, Yuji*; Hidaka, Koshi*; Tsuda, Yuko*; Kiso, Yoshiaki*
no journal, ,
HIV protease is known as a drug target protein. To compare interactions between HIVPR and inhibitors, single-chained HIVPR linked by a disulfide bond was designed as N98C mutant. Both WT N98C and A17 N98C mutants were expressed as inclusion body and refolded by dilution method. The yield of the purified enzymes was similar to that of WT. We will report the results of interactions between inhibitors and WT N98C and A17 N98C mutants by physicochemical and crystal structure analyses.
Adachi, Motoyasu; Arai, Shigeki; Tamada, Taro; Kuroki, Ryota; Hidaka, Koshi*; Kimura, Toru*; Kiso, Yoshiaki*
no journal, ,
To obtain information for design of inhibitor against drug resistant mutant HIV protease, we determined X-ray crystal structures of A17 type HIV-1 protease complexed with inhibitors of lopinavir and KNI-1657. The gene of A17 type HIV-1 protease was synthesized chemically, and prepared using E. coli expression system. The results showed that affinity of lopinavir to the protease was 700 times lower than that of wild-type, and KNI-1657 has 20 times higher affinity to the protease than lopinavir. The crystals of complex were obtained using PEG4000 as precipitant. The diffraction data were collected at PF and SPring-8, and the structures were refined at R-factor of 19%.
Hidaka, Koshi*; Adachi, Motoyasu; Kuroki, Ryota; Tokai, Satoko*; Akaji, Kenichi*; Tsuda, Yuko*; Kiso, Yoshiaki*
no journal, ,
In the HIV protease inhibitor study, we faced on a problem of much difference in activity of a compound with 2,6-dimethylphenoxyacetyl moiety against the enzyme and the virus. The protease inhibitory potency was plateau because of the limited structural modifications. Therefore, we shifted to modify the property such as reduction of the hydrophobicity. During the struggles, amino substitution succeeded in improving the water solubility to enhance the anti-HIV activity and with the sustained protease inhibitory activity. This result stimulated us to modify the Apns-containing inhibitors for the development of therapeutic drug and to utilize them as aspartic proteases probes.
Adachi, Motoyasu; Tamada, Taro; Hidaka, Koshi*; Hayashi, Yoshio*; Freire, E.*; Kiso, Yoshiaki*; Kuroki, Ryota
no journal, ,
The development of HIV protease (HIVPR) inhibitors is regarded as a major success of structure-based drug design, and the inhibitors of HIVPR are important compounds to establish highly active antiretroviral therapy for AIDS. Adverse effects linked to the use of HIVPR inhibitors, and the emergence of HIV mutants resistant to current drugs, remain critical factors in the clinical failure of antiviral therapy. We are currently working on structure analysis of HIVPR inhibitor complex using both ultra-high resolution X-ray and neutron crystallography. To obtain the HIVPR, the gene of HIVPR was expressed in E. coli and successfully refolded. A crystal of HIVPR with KNI-272 was obtained. This crystal diffracted X-ray beyond 1.1 angstroms resolution, which is much higher than the previous structure. Refinement using anisotropic temperature factors has produced more detailed structure and dynamic information about the HIVPR inhibitor complex, including details of bound water molecules.