Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Nakagawa, Hiroshi; Yonetani, Yoshiteru*; Nakajima, Kenji; Kawamura, Seiko; Kikuchi, Tatsuya*; Inamura, Yasuhiro; Kataoka, Mikio*; Kono, Hidetoshi*
JPS Conference Proceedings (Internet), 33, p.011101_1 - 011101_6, 2021/03
Hydration water dynamics were measured by quasi-elastic neutron scattering with Hn O/D
O/D O contrast for two DNA dodecamers, 5'CGCG
O contrast for two DNA dodecamers, 5'CGCG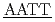 CGCG'3 and 5'CGCG
CGCG'3 and 5'CGCG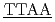 CGCG'3, which have been computationally shown to be structurally rigid and flexible, respectively. The dynamical transitions of the hydration water as well as DNA were observed for both sequences at approximately 240 K. Above the transition temperature, the mean square displacements of the hydration water for the rigid sequence were smaller than those for the flexible one. Furthermore, the relaxation time of the hydration water was longer in the rigid DNA than in the flexible DNA. We suggest that hydration water dynamics on the picosecond timescale are associated with sequence-dependent deformability of DNA.
CGCG'3, which have been computationally shown to be structurally rigid and flexible, respectively. The dynamical transitions of the hydration water as well as DNA were observed for both sequences at approximately 240 K. Above the transition temperature, the mean square displacements of the hydration water for the rigid sequence were smaller than those for the flexible one. Furthermore, the relaxation time of the hydration water was longer in the rigid DNA than in the flexible DNA. We suggest that hydration water dynamics on the picosecond timescale are associated with sequence-dependent deformability of DNA.
Ikebe, Kimiyoshi; Sakuraba, Shun*; Kono, Hidetoshi
PLOS Computational Biology, 12(3), p.e1004788_1 - e1004788_13, 2016/03
Times Cited Count:49 Percentile:90.55(Biochemical Research Methods)Kono, Hidetoshi; Shirayama, Kazuyoshi*; Arimura, Yasuhiro*; Tachiwana, Hiroaki*; Kurumizaka, Hitoshi*
PLOS ONE (Internet), 10(3), p.e0120635_1 - e0120635_15, 2015/03
Times Cited Count:28 Percentile:64.06(Multidisciplinary Sciences)Kai, Takeshi; Tokuhisa, Atsushi*; Moribayashi, Kengo; Fukuda, Yuji; Kono, Hidetoshi; Go, Nobuhiro*
Journal of the Physical Society of Japan, 83(9), p.094301_1 - 094301_5, 2014/09
Times Cited Count:1 Percentile:11.13(Physics, Multidisciplinary)no abstracts in English
Nakagawa, Hiroshi; Yonetani, Yoshiteru; Nakajima, Kenji; Kawamura, Seiko; Kikuchi, Tatsuya; Inamura, Yasuhiro; Kataoka, Mikio; Kono, Hidetoshi
Physical Review E, 90(2), p.022723_1 - 022723_11, 2014/08
Times Cited Count:10 Percentile:52.74(Physics, Fluids & Plasmas)Molecular dynamics (MD) simulations and Quasi-Elastic Neutron Scattering (QENS) experiments were conducted on hydrated two DNA dodecamers with distinct deformability; 5'CGCGAATTCGCG3' and 5'CGCGTTAACGCG3'. The former is known to be rigid and the latter to be flexible. The mean-square displacements (MSDs) of DNA dodecamers exhibit so-called dynamical transition around 200-240 K for both sequences. To investigate the DNA sequence dependent dynamics, the dynamics of DNA and hydration water above the transition temperature were examined using both MD simulations and QENS experiments. The fluctuation amplitude of the AATT central tetramer is smaller, and its relaxation time is longer, than that observed in TTAA, suggesting that the AT step is kinetically more stable than TA. The sequence-dependent local base pair step dynamics correlate with the kinetics of breaking the hydrogen bond between DNA and hydration water. The sequence dependent DNA base pair step fluctuations appear above the dynamical transition temperature. Together with these results, we conclude that DNA deformability is related to the local dynamics of base pair step, themselves coupled to hydration water in the minor groove.
Ikebe, Kimiyoshi; Sakuraba, Shun; Kono, Hidetoshi
Journal of Computational Chemistry, 35(1), p.39 - 50, 2014/01
Times Cited Count:16 Percentile:44.17(Chemistry, Multidisciplinary)Kai, Takeshi; Tokuhisa, Atsushi*; Kono, Hidetoshi
Journal of the Physical Society of Japan, 82(11), p.114301_1 - 114301_5, 2013/11
Times Cited Count:1 Percentile:10.93(Physics, Multidisciplinary)no abstracts in English
Yonetani, Yoshiteru; Kono, Hidetoshi
Journal of Physical Chemistry B, 117(25), p.7535 - 7545, 2013/06
Times Cited Count:16 Percentile:37.59(Chemistry, Physical)Ikeda, Shiro*; Kono, Hidetoshi
Tokei Suri, 61(1), p.135 - 146, 2013/06
no abstracts in English
Sunami, Tomoko; Kono, Hidetoshi
PLOS ONE (Internet), 8(2), p.e56080_1 - e56080_12, 2013/02
Times Cited Count:12 Percentile:28.73(Multidisciplinary Sciences)Kono, Hidetoshi; Ikeda, Shiro*
Hoshako, 26(1), p.38 - 43, 2013/01
A new strong, short-pulse and coherent X-ray light source, which has been built at SPring8, is expected to achieve a single molecular imaging. With this 109 times brighter light than that of the current synchrotrons, we have to measure the diffraction within a few femto-second before the molecule is destroyed due to the radiation damage. Thus, the obtained diffraction pattern will be very sparse. We have recently developed a phase retrieval method based on the Bayesian statistics that is applicable for such a sparse diffraction pattern. In this article, we introduce the method.
Yonetani, Yoshiteru; Kono, Hidetoshi
Ansanburu, 14(4), p.182 - 186, 2012/10
no abstracts in English
Yamasaki, Satoshi*; Terada, Toru*; Kono, Hidetoshi; Shimizu, Kentaro*; Sarai, Akinori*
Nucleic Acids Research, 40(17), p.e129_1 - e129_7, 2012/09
Times Cited Count:20 Percentile:44.63(Biochemistry & Molecular Biology)
Adan, J.*; Yoshii, Shuhei*; Kono, Hidetoshi; Miyata, Makoto*
Journal of Bacteriology, 194(12), p.3050 - 3057, 2012/06
Times Cited Count:10 Percentile:25.07(Microbiology)Tokuhisa, Atsushi*; Taka, Junichiro*; Kono, Hidetoshi; Go, Nobuhiro*
Acta Crystallographica Section A, 68(3), p.366 - 381, 2012/05
Times Cited Count:20 Percentile:80.81(Chemistry, Multidisciplinary)Kono, Hidetoshi; Imanishi, Miki*; Negi, Shigeru*; Tatsutani, Kazuya*; Sakaeda, Yui*; Hashimoto, Ayaka*; Nakayama, Chie*; Futaki, Shiro*; Sugiura, Yukio*
FEBS Letters, 586(6), p.918 - 923, 2012/03
Times Cited Count:5 Percentile:13.86(Biochemistry & Molecular Biology)Ikeda, Shiro*; Kono, Hidetoshi
Optics Express (Internet), 20(4), p.3375 - 3387, 2012/02
Times Cited Count:11 Percentile:48.22(Optics)Yonetani, Yoshiteru; Kono, Hidetoshi
Biophysical Chemistry, 160(1), p.54 - 61, 2012/01
Times Cited Count:21 Percentile:53.85(Biochemistry & Molecular Biology)Yonetani, Yoshiteru; Kono, Hidetoshi
Yuragi To Seitai Kino, p.66 - 71, 2010/10
no abstracts in English
Horie, Akira*; Tomita, Takeo*; Saiki, Asako*; Kono, Hidetoshi; Taka, Hikari*; Mineki, Reiko*; Fujimura, Tsutomu*; Nishiyama, Chiharu*; Kuzuyama, Tomohisa*; Nishiyama, Makoto*
Nature Chemical Biology, 5(9), p.673 - 679, 2009/09
Times Cited Count:45 Percentile:67.54(Biochemistry & Molecular Biology)