Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Tashiro, Koji*; Kusaka, Katsuhiro*; Hosoya, Takaaki*; Ohara, Takashi; Hanesaka, Makoto*; Yoshizawa, Yoshinori*; Yamamoto, Hiroko*; Niimura, Nobuo*; Tanaka, Ichiro*; Kurihara, Kazuo*; et al.
Macromolecules, 51(11), p.3911 - 3922, 2018/06
Times Cited Count:5 Percentile:17.66(Polymer Science) binding site of a
binding site of a  -lactamase from the halophile by anomalous X-ray diffraction
-lactamase from the halophile by anomalous X-ray diffractionArai, Shigeki; Shibazaki, Chie; Shimizu, Rumi; Adachi, Motoyasu; Tamada, Taro; Tokunaga, Hiroko*; Ishibashi, Matsujiro*; Tokunaga, Masao*; Kuroki, Ryota
Kyushu Shinkurotoronko Kenkyu Senta Nempo, 2014, p.17 - 19, 2016/03
no abstracts in English
 resolution
resolutionUemura, Takuya*; Kita, Akiko*; Watanabe, Yoshihiko*; Adachi, Motoyasu; Kuroki, Ryota; Morimoto, Yukio*
Biochemical and Biophysical Research Communications, 469(2), p.158 - 163, 2016/01
Times Cited Count:16 Percentile:52.34(Biochemistry & Molecular Biology)Tamada, Taro; Shinmi, Daisuke*; Ikeda, Masahiro*; Yonezawa, Yasushi*; Kataoka, Shiro*; Kuroki, Ryota; Mori, Eiji*; Motoki, Kazuhiro*
Scientific Reports (Internet), 5, p.17936_1 - 17936_12, 2015/12
Times Cited Count:25 Percentile:62.95(Multidisciplinary Sciences)The fully human monoclonal antibody KMTR2 acts as a strong direct agonist for tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) receptor 2 (TRAIL-R2), which is capable of inducing apoptotic cell death without cross-linking. To investigate the mechanism of direct agonistic activity induced by KMTR2, the crystal structure of the extracellular region of TRAIL-R2 and a Fab fragment derived from KMTR2 (KMTR2-Fab) was determined to 2.1  resolution. Two KMTR2-Fabs assembled with the complementarity-determining region 2 of the light chain via two-fold crystallographic symmetry, suggesting that the KMTR2-Fab assembly tended to enhance TRAIL-R2 oligomerization. A single mutation at Asn53 to Arg located at the two-fold interface in the KMTR2 resulted in a loss of its apoptotic activity, although it retained its antigen-binding activity. These results indicate that the strong agonistic activity, such as apoptotic signaling and tumor regression, induced by KMTR2 is attributed to TRAIL-R2 superoligomerization induced by the interdimerization of KMTR2.
resolution. Two KMTR2-Fabs assembled with the complementarity-determining region 2 of the light chain via two-fold crystallographic symmetry, suggesting that the KMTR2-Fab assembly tended to enhance TRAIL-R2 oligomerization. A single mutation at Asn53 to Arg located at the two-fold interface in the KMTR2 resulted in a loss of its apoptotic activity, although it retained its antigen-binding activity. These results indicate that the strong agonistic activity, such as apoptotic signaling and tumor regression, induced by KMTR2 is attributed to TRAIL-R2 superoligomerization induced by the interdimerization of KMTR2.
Hiromoto, Takeshi; Adachi, Motoyasu; Shibazaki, Chie; Schrader, T. E.*; Ostermann, A.*; Kuroki, Ryota
JPS Conference Proceedings (Internet), 8, p.033003_1 - 033003_6, 2015/09
T4 phage lysozyme (T4L) is an endoacetylmuramidase that degrades the murein of the bacterial cell wall by cleaving the  -1,4-glycosidic bond between
-1,4-glycosidic bond between 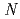 -acetylmuramic acid and
-acetylmuramic acid and 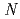 -acetylglucosamine. We previously reported that the substitution of the catalytic Thr26 with the nucleophilic His converts the wild-type (WT) T4L from an inverting to a retaining glycosidase, in which the
-acetylglucosamine. We previously reported that the substitution of the catalytic Thr26 with the nucleophilic His converts the wild-type (WT) T4L from an inverting to a retaining glycosidase, in which the  -configuration of the substrate is retained in the product. We also found that the Thr26His (T26H) mutant can catalyze a transglycosylation reaction more effectively than hydrolysis, although the WT-T4L has no transglycosidase activity. To clarify the role of the substituted His26 in transglycosylation and to investigate its relationship to the neighboring acidic residue Asp20 using neutron crystallography, a perdeuterated recombinant protein of the T26H mutant (d-T26H) was prepared for crystallization. The perdeuterated form was produced in
-configuration of the substrate is retained in the product. We also found that the Thr26His (T26H) mutant can catalyze a transglycosylation reaction more effectively than hydrolysis, although the WT-T4L has no transglycosidase activity. To clarify the role of the substituted His26 in transglycosylation and to investigate its relationship to the neighboring acidic residue Asp20 using neutron crystallography, a perdeuterated recombinant protein of the T26H mutant (d-T26H) was prepared for crystallization. The perdeuterated form was produced in 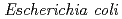 cells cultured in deuterated-rich media. After purification, the d-T26H mutant was crystallized under deuterated conditions and grown to a volume of 0.12 mm
cells cultured in deuterated-rich media. After purification, the d-T26H mutant was crystallized under deuterated conditions and grown to a volume of 0.12 mm using a macroseeding technique. A preliminary neutron-diffraction experiment at 100 K at the FRM II research reactor (Munich, Germany) gave diffraction spots of up to 2.8
using a macroseeding technique. A preliminary neutron-diffraction experiment at 100 K at the FRM II research reactor (Munich, Germany) gave diffraction spots of up to 2.8  resolution after a 1.5 h exposure.
resolution after a 1.5 h exposure.
Tomoyori, Katsuaki; Kurihara, Kazuo; Tamada, Taro; Kuroki, Ryota
JPS Conference Proceedings (Internet), 8, p.036004_1 - 036004_6, 2015/09
We aim to build a high-resolution neutron time-of-flight diffractometer for biomacromolecules at the Materials and Life Science Experimental Facility (MLF) at the Japan Proton Accelerator Research Complex (J-PARC) that allows the collection of neutron diffraction data from crystals with unit cells of 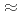 250
250  ;. Considering both the flux and pulse width necessary to realize data collection covering a minimum d-spacing of 2.0
;. Considering both the flux and pulse width necessary to realize data collection covering a minimum d-spacing of 2.0  ; and with a unit cell constant of
; and with a unit cell constant of 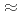 250
250  ; we chose a decoupled moderator (DM) as the appropriate source for this high-resolution diffractometer. We considered a simple instrumentation model that includes a moderator, neutron guide, sample size, and neutron detector; we then investigated its spot separation performance and estimated the instrumental parameters for the design of a new diffractometer for protein crystals with large unit cells at J-PARC/MLF. It is preferable to extend the total flight path to resolve Bragg reflections for protein crystals with large unit cells as the scattering angle increases. Meanwhile, to ensure resolvable detection capacity at the middle scattering angle region (2
; we chose a decoupled moderator (DM) as the appropriate source for this high-resolution diffractometer. We considered a simple instrumentation model that includes a moderator, neutron guide, sample size, and neutron detector; we then investigated its spot separation performance and estimated the instrumental parameters for the design of a new diffractometer for protein crystals with large unit cells at J-PARC/MLF. It is preferable to extend the total flight path to resolve Bragg reflections for protein crystals with large unit cells as the scattering angle increases. Meanwhile, to ensure resolvable detection capacity at the middle scattering angle region (2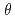
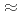 90
90 ), it is necessary to restrict the angular divergence. In the case of
), it is necessary to restrict the angular divergence. In the case of 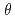

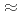 0.2
0.2 , scattering angles from around 2
, scattering angles from around 2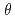
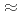 90
90 to higher backscattering angles are more efficient for protein crystals with large unit cells (
to higher backscattering angles are more efficient for protein crystals with large unit cells (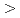 250
250  ) with a resolution of 2.0
) with a resolution of 2.0  .
.
 sp. AS-131 isolated from Antarctic Ocean
sp. AS-131 isolated from Antarctic OceanYonezawa, Yasushi*; Nagayama, Aiko*; Tokunaga, Hiroko*; Ishibashi, Matsujiro*; Arai, Shigeki; Kuroki, Ryota; Watanabe, Keiichi*; Arakawa, Tsutomu*; Tokunaga, Masao*
Protein Journal, 34(4), p.275 - 283, 2015/08
Times Cited Count:4 Percentile:10.94(Biochemistry & Molecular Biology)Nucleoside diphosphate kinase isolated from psychrophilic  sp. AS-131 (ASNDK) was expressed in
sp. AS-131 (ASNDK) was expressed in  and purified to homogeneity. Comparing to mesophilic NDK isolated from
and purified to homogeneity. Comparing to mesophilic NDK isolated from  , ASNDK exhibited highly elevated thermolability: (1)
, ASNDK exhibited highly elevated thermolability: (1) 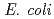 expression at 37
expression at 37 C as a denatured insoluble form, and (2) 30
C as a denatured insoluble form, and (2) 30 C lower optimum temperature of enzymatic activity. The subunit structure of ASNDK was suggested to be dimer, as in NDKs isolated from moderate halophiles.
C lower optimum temperature of enzymatic activity. The subunit structure of ASNDK was suggested to be dimer, as in NDKs isolated from moderate halophiles.
Unno, Masayoshi*; Ishikawa, Kumiko*; Kusaka, Katsuhiro*; Tamada, Taro; Hagiwara, Yoshinori*; Sugishima, Masakazu*; Wada, Kei*; Yamada, Taro*; Tomoyori, Katsuaki; Hosoya, Takaaki*; et al.
Journal of the American Chemical Society, 137(16), p.5452 - 5460, 2015/04
Times Cited Count:28 Percentile:63.97(Chemistry, Multidisciplinary)Phycocyanobilin, a light-harvesting and photoreceptor pigment in higher plants, algae, and cyanobacteria, is synthesized from biliverdin IX (BV) by phycocyanobilin:ferredoxin oxidoreductase (PcyA) via two steps of two-proton-coupled two-electron reduction. We determined the neutron structure of PcyA from cyanobacteria complexed with BV, revealing the exact location of the hydrogen atoms involved in catalysis. Notably, approximately half of the BV bound to PcyA was BVH
(BV) by phycocyanobilin:ferredoxin oxidoreductase (PcyA) via two steps of two-proton-coupled two-electron reduction. We determined the neutron structure of PcyA from cyanobacteria complexed with BV, revealing the exact location of the hydrogen atoms involved in catalysis. Notably, approximately half of the BV bound to PcyA was BVH , a state in which all four pyrrole nitrogen atoms were protonated. The protonation states of BV complemented the protonation of adjacent Asp105. The "axial "water molecule that interacts with the neutral pyrrole nitrogen of the A-ring was identified. His88 N
, a state in which all four pyrrole nitrogen atoms were protonated. The protonation states of BV complemented the protonation of adjacent Asp105. The "axial "water molecule that interacts with the neutral pyrrole nitrogen of the A-ring was identified. His88 N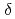 was protonated to form a hydrogen bond with the lactam O atom of the BV A-ring. His88 and His74 were linked by hydrogen bonds via H
was protonated to form a hydrogen bond with the lactam O atom of the BV A-ring. His88 and His74 were linked by hydrogen bonds via H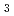 O
O . These results imply that Asp105, His88, and the axial water molecule contribute to proton transfer during PcyA catalysis.
. These results imply that Asp105, His88, and the axial water molecule contribute to proton transfer during PcyA catalysis.
 -lactamase from the moderate halophile
-lactamase from the moderate halophile  sp.560 and the discovery of a Cs
sp.560 and the discovery of a Cs -selective binding site
-selective binding siteArai, Shigeki; Yonezawa, Yasushi*; Okazaki, Nobuo*; Matsumoto, Fumiko*; Shibazaki, Chie; Shimizu, Rumi; Yamada, Mitsugu*; Adachi, Motoyasu; Tamada, Taro; Kawamoto, Masahide*; et al.
Acta Crystallographica Section D, 71(3), p.541 - 554, 2015/03
Times Cited Count:7 Percentile:50.44(Biochemical Research Methods)The crystal structure of halophilic  -lactamase from
-lactamase from  sp.560 (HaBLA) was determined using X-ray crystallography. Moreover, the locations of bound Sr
sp.560 (HaBLA) was determined using X-ray crystallography. Moreover, the locations of bound Sr and Cs
and Cs ions were identified by anomalous X-ray diffraction. The location of one Cs
ions were identified by anomalous X-ray diffraction. The location of one Cs specific binding site was identified on HaBLA even in the presence of 9-fold molar excess of Na
specific binding site was identified on HaBLA even in the presence of 9-fold molar excess of Na (90 mM Na
(90 mM Na /10 mM Cs
/10 mM Cs ). This Cs
). This Cs binding site is formed by two main-chain O atoms and an aromatic ring of a side chain of Trp. An aromatic ring of Trp interacts with Cs
binding site is formed by two main-chain O atoms and an aromatic ring of a side chain of Trp. An aromatic ring of Trp interacts with Cs by the cation-
by the cation-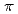 interaction. The observation of a selective and high-affinity Cs
interaction. The observation of a selective and high-affinity Cs binding site provides important information that is useful for designing artificial Cs
binding site provides important information that is useful for designing artificial Cs binding sites useful in bioremediation of radioactive isotopes.
binding sites useful in bioremediation of radioactive isotopes.
 -glucosyltransferase from
-glucosyltransferase from 
Hiromoto, Takeshi; Honjo, Eijiro*; Noda, Hisanobu*; Tamada, Taro; Kazuma, Kohei*; Suzuki, Masahiko*; Blaber, M.; Kuroki, Ryota
Protein Science, 24(3), p.395 - 407, 2015/03
Times Cited Count:62 Percentile:88.89(Biochemistry & Molecular Biology)UDP-glucose: anthocyanidin 3- -glucosyltransferase (UGT78K6) from
-glucosyltransferase (UGT78K6) from  catalyzes the transfer of glucose from UDP-glucose to anthocyanidins such as delphinidin. To understand the acceptor-recognition scheme of UGT78K6, the crystal structure of UGT78K6 and its complex forms with anthocyanidin delphinidin and petunidin, and flavonol kaempferol were determined to resolutions of 1.85
catalyzes the transfer of glucose from UDP-glucose to anthocyanidins such as delphinidin. To understand the acceptor-recognition scheme of UGT78K6, the crystal structure of UGT78K6 and its complex forms with anthocyanidin delphinidin and petunidin, and flavonol kaempferol were determined to resolutions of 1.85  , 2.55
, 2.55  , 2.70
, 2.70  and 1.75
and 1.75  respectively. The anthocyanidin- and flavonol-acceptor binding details are almost identical in each complex structure, although the glucosylation activities against each acceptor were significantly different. The acceptor substrates in UGT78K6 are reversely bound to its binding site by a 180
respectively. The anthocyanidin- and flavonol-acceptor binding details are almost identical in each complex structure, although the glucosylation activities against each acceptor were significantly different. The acceptor substrates in UGT78K6 are reversely bound to its binding site by a 180 rotation about the O1-O3 axis of the flavonoid backbones observed in
rotation about the O1-O3 axis of the flavonoid backbones observed in 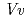 GT1 and UGT78G1. These substrate recognition schemes suggest the potential for controlled synthesis of natural pigments.
GT1 and UGT78G1. These substrate recognition schemes suggest the potential for controlled synthesis of natural pigments.
Kuroki, Ryota; Miyazaki, Hiroshi*; Kato, Takashi
Ketsueki Furonteia, 25(2), p.171 - 180, 2015/02
no abstracts in English
Tashiro, Koji*; Hanesaka, Makoto*; Yamamoto, Hiroko*; Wasanasuk, K.*; Jayaratri, P.*; Yoshizawa, Yoshinori*; Tanaka, Ichiro*; Niimura, Nobuo*; Kusaka, Katsuhiro*; Hosoya, Takaaki*; et al.
Kobunshi Rombunshu, 71(11), p.508 - 526, 2014/11
Times Cited Count:6 Percentile:22.16(Polymer Science)The crystal structure analysis of various polymer substances has been reviewed on the basis of wide-angle high-energy X-ray and neutron diffraction data. The progress in structural analytical techniques of polymer crystals have been reviewed at first. The structural models proposed so far were reinvestigated and new models have been proposed for various kinds of polymer crystals including polyethylene, poly(vinyl alcohol), poly(lactic acid) and its stereocomplex etc. The hydrogen atomic positions were also clarified by the quantitative analysis of wide-angle neutron diffraction data, from which the physical properties of polymer crystals have been evaluated theoretically. The bonded electron density distribution has been estimated for a polydiacetylene single crystal on the basis of the so-called X-N method or by the combination of structural information derived from X-ray and neutron diffraction data analysis. Some comments have been added about future developments in the field of structure-property relationship determination.

Adachi, Motoyasu; Hirayama, Hiroshi; Shimizu, Rumi; Sato, Katsuya; Narumi, Issey*; Kuroki, Ryota
Protein Science, 23(10), p.1349 - 1358, 2014/10
Times Cited Count:9 Percentile:25.18(Biochemistry & Molecular Biology)Pleiotropic protein promoting DNA repair A (PprA) is a key protein that facilitates the extreme radioresistance of  . To clarify the role of PprA in the radioresistance mechanism, the interaction between recombinant PprA expressed in Escherichia coli with several double-stranded DNAs was investigated. In a gel-shift assay, the band shift of supercoiled pUC19 DNA caused by the binding of PprA showed a bimodal distribution, which was promoted by the addition of 1 mM Mg, Ca, or Sr ions. The dissociation constant of the PprA-supercoiled pUC19 DNA complex, calculated from the relative portions of shifted bands, was 0.6
. To clarify the role of PprA in the radioresistance mechanism, the interaction between recombinant PprA expressed in Escherichia coli with several double-stranded DNAs was investigated. In a gel-shift assay, the band shift of supercoiled pUC19 DNA caused by the binding of PprA showed a bimodal distribution, which was promoted by the addition of 1 mM Mg, Ca, or Sr ions. The dissociation constant of the PprA-supercoiled pUC19 DNA complex, calculated from the relative portions of shifted bands, was 0.6 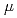 M with a Hill coefficient of 3.3 in the presence of 1 mM Mg acetate. This indicates that at least 281 PprA molecules are required to saturate a supercoiled pUC19 DNA, which is consistent with the number of bound PprA molecules estimated by the UV absorption of the PprA-pUC19 complex purified by gel filtration. This saturation also suggests linear polymerization of PprA along the dsDNA. On the other hand, the bands of linear dsDNA and nicked circular dsDNA that eventually formed PprA complexes did not saturate, but created larger molecular complexes when the PprA concentration was greater than 1.3
M with a Hill coefficient of 3.3 in the presence of 1 mM Mg acetate. This indicates that at least 281 PprA molecules are required to saturate a supercoiled pUC19 DNA, which is consistent with the number of bound PprA molecules estimated by the UV absorption of the PprA-pUC19 complex purified by gel filtration. This saturation also suggests linear polymerization of PprA along the dsDNA. On the other hand, the bands of linear dsDNA and nicked circular dsDNA that eventually formed PprA complexes did not saturate, but created larger molecular complexes when the PprA concentration was greater than 1.3 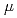 M. This result implies that DNA-bound PprA aids association of the termini of damaged DNAs, which is regulated by the concentration of PprA.
M. This result implies that DNA-bound PprA aids association of the termini of damaged DNAs, which is regulated by the concentration of PprA.
Kuroki, Ryota
Shinka O Tsuzukeru Kozo Seibutsugaku; Aratana Tampakushitsu Kino No Kaimei To Soshutsu, p.93 - 109, 2014/09
no abstracts in English
Kuroki, Ryota
Nihon No Kesshogaku, 2; Sono Kagayakashii Hatten, P. 392, 2014/07
no abstracts in English
 sp.593
sp.593Arai, Shigeki; Yonezawa, Yasushi*; Ishibashi, Matsujiro*; Matsumoto, Fumiko*; Adachi, Motoyasu; Tamada, Taro; Tokunaga, Hiroko*; Blaber, M.; Tokunaga, Masao*; Kuroki, Ryota
Acta Crystallographica Section D, 70(3), p.811 - 820, 2014/03
Times Cited Count:12 Percentile:62.81(Biochemical Research Methods)In order to clarify the structural basis of halophilic characteristics of an alkaline phosphatase derived from the moderate halophile  sp.593 (HaAP), the tertiary structure of HaAP was determined to 2.1
sp.593 (HaAP), the tertiary structure of HaAP was determined to 2.1 resolution by X-ray crystallography. Structural properties of surface negative charge and core hydrophobicity are shown to be intermediate between halophile and non-halophile characteristics, and may explain the unique functional adaptation to a wide-range of salt concentration.
resolution by X-ray crystallography. Structural properties of surface negative charge and core hydrophobicity are shown to be intermediate between halophile and non-halophile characteristics, and may explain the unique functional adaptation to a wide-range of salt concentration.
Adachi, Motoyasu; Arai, Shigeki; Hiromoto, Takeshi; Kuroki, Ryota
Hamon, 24(1), p.45 - 49, 2014/02
Protein structure analysis using neutron diffraction (neutron protein crystallography; NPC) is gaining greater importance in the understanding of structure and function relationships of biological macromolecules such as proteins and DNA. Current developments of neutron diffractometers installed at the JAEA research reactor and pulsed neutron source permit observation of the locations of hydrogen atoms and hydrating water molecules and help understanding of important mechanisms of chemical reactions catalyzed by biological macromolecules. Here, we introduce practical approaches of NPC including sample preparation, crystal growth, structure determination and utilization of information obtained from NPC.
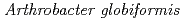 ; Assignment of bound diatomic molecules as O
; Assignment of bound diatomic molecules as O
Murakawa, Takeshi*; Hayashi, Hideyuki*; Sunami, Tomoko; Kurihara, Kazuo; Tamada, Taro; Kuroki, Ryota; Suzuki, Mamoru*; Tanizawa, Katsuyuki*; Okajima, Toshihide*
Acta Crystallographica Section D, 69(12), p.2483 - 2494, 2013/12
Times Cited Count:14 Percentile:68.53(Biochemical Research Methods)The crystal structure of a Cu amine oxidase from 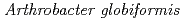 was determined at 1.08
was determined at 1.08  resolution with the use of low-molecular-weight polyethylene glycol (LMW PEG; average molecular weight
resolution with the use of low-molecular-weight polyethylene glycol (LMW PEG; average molecular weight  200) as a cryoprotectant. The final crystallographic
200) as a cryoprotectant. The final crystallographic 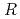 -factor and
-factor and 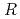
 value are 13.0% and 15.0%, respectively. Several molecules of LMW PEG were found to occupy cavities in the protein interior including the active site, which resulted in the marked reduction of the overall
value are 13.0% and 15.0%, respectively. Several molecules of LMW PEG were found to occupy cavities in the protein interior including the active site, which resulted in the marked reduction of the overall 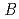 factor and consequently led to a sub-atomic resolution structure for a relatively large protein with a monomer molecular weight of
factor and consequently led to a sub-atomic resolution structure for a relatively large protein with a monomer molecular weight of  70,000. About 40% of all the presumed hydrogen atoms were observed as clear electron densities in the
70,000. About 40% of all the presumed hydrogen atoms were observed as clear electron densities in the 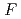
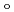 -
- 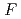
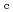 difference map. Multiple minor conformers were also identified for many residues. Anisotropic displacement fluctuations were evaluated in the active site that contains a post-translationally derived quinone cofactor and a Cu atom. Furthermore, diatomic molecules, most likely molecular oxygen, are bound to the protein, one of which is located in the region that has been previously proposed as an entry route for the substrate dioxygen from the central cavity of the dimer interface to the active site.
difference map. Multiple minor conformers were also identified for many residues. Anisotropic displacement fluctuations were evaluated in the active site that contains a post-translationally derived quinone cofactor and a Cu atom. Furthermore, diatomic molecules, most likely molecular oxygen, are bound to the protein, one of which is located in the region that has been previously proposed as an entry route for the substrate dioxygen from the central cavity of the dimer interface to the active site.
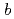
 reductase by X-ray crystallography; New insights into regulation of efficient electron transfer
reductase by X-ray crystallography; New insights into regulation of efficient electron transferYamada, Mitsugu*; Tamada, Taro; Takeda, Kazuki*; Matsumoto, Fumiko*; Ono, Hiraku*; Kosugi, Masayuki*; Takaba, Kiyofumi*; Shoyama, Yoshinari*; Kimura, Shigenobu*; Kuroki, Ryota; et al.
Journal of Molecular Biology, 425(22), p.4295 - 4306, 2013/11
Times Cited Count:21 Percentile:50.33(Biochemistry & Molecular Biology)NADH-Cytochrome 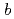
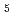 reductase (b5R), a flavoprotein consisting of NADH and flavin adenine dinucleotide (FAD) binding domains, catalyzes electron transfer from the two-electron carrier NADH to the one-electron carrier cytochrome
reductase (b5R), a flavoprotein consisting of NADH and flavin adenine dinucleotide (FAD) binding domains, catalyzes electron transfer from the two-electron carrier NADH to the one-electron carrier cytochrome 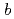
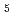 (Cb5). The crystal structures of both the fully reduced form and the oxidized form of porcine liver b5R were determined. In the reduced b5R structure determined at 1.68
(Cb5). The crystal structures of both the fully reduced form and the oxidized form of porcine liver b5R were determined. In the reduced b5R structure determined at 1.68 resolution, the relative configuration of the two domains was slightly shifted in comparison with that of the oxidized form. This shift resulted in an increase in the solvent-accessible surface area of FAD and created a new hydrogen-bonding interaction between the N5 atom of the isoalloxazine ring of FAD and the hydroxyl oxygen atom of Thr66, which is considered to be a key residue in the release of a proton from the N5 atom. The isoalloxazine ring of FAD in the reduced form is flat as in the oxidized form and stacked together with the nicotinamide ring of NAD
resolution, the relative configuration of the two domains was slightly shifted in comparison with that of the oxidized form. This shift resulted in an increase in the solvent-accessible surface area of FAD and created a new hydrogen-bonding interaction between the N5 atom of the isoalloxazine ring of FAD and the hydroxyl oxygen atom of Thr66, which is considered to be a key residue in the release of a proton from the N5 atom. The isoalloxazine ring of FAD in the reduced form is flat as in the oxidized form and stacked together with the nicotinamide ring of NAD . Determination of the oxidized b5R structure, including the hydrogen atoms, determined at 0.78
. Determination of the oxidized b5R structure, including the hydrogen atoms, determined at 0.78 resolution revealed the details of a hydrogen-bonding network from the N5 atom of FAD to His49 via Thr66. Both of the reduced and oxidized b5R structures explain how backflow in this catalytic cycle is prevented and the transfer of electrons to one-electron acceptors such as Cb5 is accelerated. Furthermore, crystallographic analysis by the cryo-trapping method suggests that re-oxidation follows a two-step mechanism. These results provide structural insights into the catalytic cycle of b5R.
resolution revealed the details of a hydrogen-bonding network from the N5 atom of FAD to His49 via Thr66. Both of the reduced and oxidized b5R structures explain how backflow in this catalytic cycle is prevented and the transfer of electrons to one-electron acceptors such as Cb5 is accelerated. Furthermore, crystallographic analysis by the cryo-trapping method suggests that re-oxidation follows a two-step mechanism. These results provide structural insights into the catalytic cycle of b5R.
 -glucosyltransferase from
-glucosyltransferase from 
Hiromoto, Takeshi; Honjo, Eijiro*; Tamada, Taro; Noda, Hisanobu*; Kazuma, Kohei*; Suzuki, Masahiko*; Kuroki, Ryota
Journal of Synchrotron Radiation, 20(6), p.894 - 898, 2013/11
Times Cited Count:38 Percentile:86.36(Instruments & Instrumentation)Flowers of the butterfly pea ( ) accumulate a group of polyacylated anthocyanins, named ternatins, in their petals. The first step in ternatin biosynthesis is the transfer of glucose from UDP-glucose to anthocyanidins such as delphinidin, a reaction catalyzed in
) accumulate a group of polyacylated anthocyanins, named ternatins, in their petals. The first step in ternatin biosynthesis is the transfer of glucose from UDP-glucose to anthocyanidins such as delphinidin, a reaction catalyzed in  by UDP-glucose:anthocyanidin 3-
by UDP-glucose:anthocyanidin 3- -glucosyltransferase (
-glucosyltransferase ( 3GT-A; AB185904). To elucidate the structure-function relationship of
3GT-A; AB185904). To elucidate the structure-function relationship of  3GT-A, recombinant
3GT-A, recombinant  3GT-A was expressed in
3GT-A was expressed in 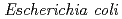 and its tertiary structure was determined to 1.85
and its tertiary structure was determined to 1.85  , resolution by using X-ray crystallography. The structure of
, resolution by using X-ray crystallography. The structure of  3GT-A shows a common folding topology, the GT-B fold, comprised of two Rossmann-like
3GT-A shows a common folding topology, the GT-B fold, comprised of two Rossmann-like  /
/ /
/ domains and a cleft located between the N- and C-domains containing two cavities that are used as binding sites for the donor (UDP-Glc) and acceptor substrates. By comparing the structure of
domains and a cleft located between the N- and C-domains containing two cavities that are used as binding sites for the donor (UDP-Glc) and acceptor substrates. By comparing the structure of  3GT-A with that of the flavonoid glycosyltransferase
3GT-A with that of the flavonoid glycosyltransferase 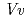 GT1 from red grape (
GT1 from red grape (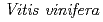 ) in complex with UDP-2-deoxy-2-fluoro glucose and kaempferol, locations of the catalytic His-Asp dyad and the residues involved in recognizing UDP-2-deoxy-2-fluoro glucose were essentially identical in
) in complex with UDP-2-deoxy-2-fluoro glucose and kaempferol, locations of the catalytic His-Asp dyad and the residues involved in recognizing UDP-2-deoxy-2-fluoro glucose were essentially identical in  3GT-A, but certain residues of
3GT-A, but certain residues of 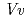 GT1 involved in binding kaempferol were found to be substituted in
GT1 involved in binding kaempferol were found to be substituted in  3GT-A. These findings are important for understanding the differentiation of acceptor-substrate recognition in these two enzymes.
3GT-A. These findings are important for understanding the differentiation of acceptor-substrate recognition in these two enzymes.