Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
 binding site of a
binding site of a  -lactamase from the halophile by anomalous X-ray diffraction
-lactamase from the halophile by anomalous X-ray diffractionArai, Shigeki; Shibazaki, Chie; Shimizu, Rumi; Adachi, Motoyasu; Tamada, Taro; Tokunaga, Hiroko*; Ishibashi, Matsujiro*; Tokunaga, Masao*; Kuroki, Ryota
Kyushu Shinkurotoronko Kenkyu Senta Nempo, 2014, p.17 - 19, 2016/03
no abstracts in English
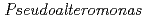 sp. AS-131 isolated from Antarctic Ocean
sp. AS-131 isolated from Antarctic OceanYonezawa, Yasushi*; Nagayama, Aiko*; Tokunaga, Hiroko*; Ishibashi, Matsujiro*; Arai, Shigeki; Kuroki, Ryota; Watanabe, Keiichi*; Arakawa, Tsutomu*; Tokunaga, Masao*
Protein Journal, 34(4), p.275 - 283, 2015/08
Times Cited Count:4 Percentile:9.94(Biochemistry & Molecular Biology)Nucleoside diphosphate kinase isolated from psychrophilic 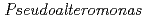 sp. AS-131 (ASNDK) was expressed in
sp. AS-131 (ASNDK) was expressed in 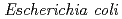 and purified to homogeneity. Comparing to mesophilic NDK isolated from
and purified to homogeneity. Comparing to mesophilic NDK isolated from 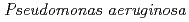 , ASNDK exhibited highly elevated thermolability: (1)
, ASNDK exhibited highly elevated thermolability: (1) 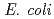 expression at 37
expression at 37 C as a denatured insoluble form, and (2) 30
C as a denatured insoluble form, and (2) 30 C lower optimum temperature of enzymatic activity. The subunit structure of ASNDK was suggested to be dimer, as in NDKs isolated from moderate halophiles.
C lower optimum temperature of enzymatic activity. The subunit structure of ASNDK was suggested to be dimer, as in NDKs isolated from moderate halophiles.
 -lactamase from the moderate halophile
-lactamase from the moderate halophile  sp.560 and the discovery of a Cs
sp.560 and the discovery of a Cs -selective binding site
-selective binding siteArai, Shigeki; Yonezawa, Yasushi*; Okazaki, Nobuo*; Matsumoto, Fumiko*; Shibazaki, Chie; Shimizu, Rumi; Yamada, Mitsugu*; Adachi, Motoyasu; Tamada, Taro; Kawamoto, Masahide*; et al.
Acta Crystallographica Section D, 71(3), p.541 - 554, 2015/03
Times Cited Count:8 Percentile:52.42(Biochemical Research Methods)The crystal structure of halophilic  -lactamase from
-lactamase from  sp.560 (HaBLA) was determined using X-ray crystallography. Moreover, the locations of bound Sr
sp.560 (HaBLA) was determined using X-ray crystallography. Moreover, the locations of bound Sr and Cs
and Cs ions were identified by anomalous X-ray diffraction. The location of one Cs
ions were identified by anomalous X-ray diffraction. The location of one Cs specific binding site was identified on HaBLA even in the presence of 9-fold molar excess of Na
specific binding site was identified on HaBLA even in the presence of 9-fold molar excess of Na (90 mM Na
(90 mM Na /10 mM Cs
/10 mM Cs ). This Cs
). This Cs binding site is formed by two main-chain O atoms and an aromatic ring of a side chain of Trp. An aromatic ring of Trp interacts with Cs
binding site is formed by two main-chain O atoms and an aromatic ring of a side chain of Trp. An aromatic ring of Trp interacts with Cs by the cation-
by the cation-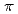 interaction. The observation of a selective and high-affinity Cs
interaction. The observation of a selective and high-affinity Cs binding site provides important information that is useful for designing artificial Cs
binding site provides important information that is useful for designing artificial Cs binding sites useful in bioremediation of radioactive isotopes.
binding sites useful in bioremediation of radioactive isotopes.
 sp.593
sp.593Arai, Shigeki; Yonezawa, Yasushi*; Ishibashi, Matsujiro*; Matsumoto, Fumiko*; Adachi, Motoyasu; Tamada, Taro; Tokunaga, Hiroko*; Blaber, M.; Tokunaga, Masao*; Kuroki, Ryota
Acta Crystallographica Section D, 70(3), p.811 - 820, 2014/03
Times Cited Count:13 Percentile:63.41(Biochemical Research Methods)In order to clarify the structural basis of halophilic characteristics of an alkaline phosphatase derived from the moderate halophile  sp.593 (HaAP), the tertiary structure of HaAP was determined to 2.1
sp.593 (HaAP), the tertiary structure of HaAP was determined to 2.1 resolution by X-ray crystallography. Structural properties of surface negative charge and core hydrophobicity are shown to be intermediate between halophile and non-halophile characteristics, and may explain the unique functional adaptation to a wide-range of salt concentration.
resolution by X-ray crystallography. Structural properties of surface negative charge and core hydrophobicity are shown to be intermediate between halophile and non-halophile characteristics, and may explain the unique functional adaptation to a wide-range of salt concentration.
Adachi, Motoyasu; Arai, Shigeki; Hiromoto, Takeshi; Kuroki, Ryota
Hamon, 24(1), p.45 - 49, 2014/02
Protein structure analysis using neutron diffraction (neutron protein crystallography; NPC) is gaining greater importance in the understanding of structure and function relationships of biological macromolecules such as proteins and DNA. Current developments of neutron diffractometers installed at the JAEA research reactor and pulsed neutron source permit observation of the locations of hydrogen atoms and hydrating water molecules and help understanding of important mechanisms of chemical reactions catalyzed by biological macromolecules. Here, we introduce practical approaches of NPC including sample preparation, crystal growth, structure determination and utilization of information obtained from NPC.
Saegusa, Jun; Kurikami, Hiroshi; Yasuda, Ryo; Kurihara, Kazuo; Arai, Shigeki; Kuroki, Ryota; Matsuhashi, Shimpei; Ozawa, Takashi; Goto, Hiroaki; Takano, Takao; et al.
Health Physics, 104(3), p.243 - 250, 2013/03
Times Cited Count:3 Percentile:24.16(Environmental Sciences)After the Nuclear accident on March 2011, water discharge from many outdoor swimming pools in the Fukushima prefecture was suspended out of concern that radiocesium in the pool water would flow into farmlands. We have reviewed the existing flocculation method for decontaminating pool water and established a practical decontamination method by demonstrating the process at several pools in the Fukushima prefecture.
Niwa, Hideharu*; Saito, Makoto*; Kobayashi, Masaki*; Harada, Yoshihisa*; Oshima, Masaharu*; Moriya, Shogo*; Matsubayashi, Katsuyuki*; Nabae, Yuta*; Kuroki, Shigeki*; Ikeda, Takashi; et al.
Journal of Power Sources, 223, p.30 - 35, 2013/02
Times Cited Count:18 Percentile:48.58(Chemistry, Physical)To design non-platinum, inexpensive, but high performance carbon-based cathode catalysts for polymer electrolyte fuel cells, it is important to elucidate the active site for oxygen reduction reaction (ORR). However, it is difficult to directly identify the active site by applying conventional structural or electronic probes to such complex systems. Here, we used C 1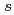 X-ray absorption spectroscopy (XAS) to observe electronic structure of carbon in iron phthalocyanine-based catalysts, and found a signature of edge exposure below the
X-ray absorption spectroscopy (XAS) to observe electronic structure of carbon in iron phthalocyanine-based catalysts, and found a signature of edge exposure below the 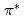 edge, whose intensity is well correlated with the ORR activity. These results demonstrate that C 1
edge, whose intensity is well correlated with the ORR activity. These results demonstrate that C 1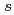 XAS can be used to characterize the ORR activity of carbon-based cathode catalysts in terms of the edge exposure.
XAS can be used to characterize the ORR activity of carbon-based cathode catalysts in terms of the edge exposure.
Kiuchi, Hisao*; Niwa, Hideharu*; Kobayashi, Masaki*; Harada, Yoshihisa*; Oshima, Masaharu*; Chokai, Masayuki*; Nabae, Yuta*; Kuroki, Shigeki*; Kakimoto, Masaaki*; Ikeda, Takashi; et al.
Electrochimica Acta, 82(1), p.291 - 295, 2012/10
Times Cited Count:14 Percentile:32.62(Electrochemistry)We study the characteristics of oxygen adsorption on metal-free carbon-based cathode catalysts derived from nitrogen-containing polyamide (PA) and nitrogen-free phenolic resin (PhRs). Electrochemical analysis and Raman spectroscopy showed higher 2-electron oxygen reduction reaction (ORR) activity and more defect sites in PA than PhRs. The increase in the amount of adsorbed oxygen in PA was also identified by oxygen adsorption isotherms. 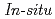 X-ray photoelectron spectroscopy reveals that graphite-like nitrogen contributes to oxygen adsorption and C=O components are dominant in PA. These experimental results indicate that the adsorbed C=O components near the graphite-like nitrogen can be assigned as active sites for 2-electron ORR.
X-ray photoelectron spectroscopy reveals that graphite-like nitrogen contributes to oxygen adsorption and C=O components are dominant in PA. These experimental results indicate that the adsorbed C=O components near the graphite-like nitrogen can be assigned as active sites for 2-electron ORR.
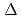 1-tetrahydrocannabinolic acid (THCA) synthase, the enzyme controlling the psychoactivity of
1-tetrahydrocannabinolic acid (THCA) synthase, the enzyme controlling the psychoactivity of 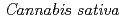
Shoyama, Yoshinari*; Tamada, Taro; Kurihara, Kazuo; Takeuchi, Ayako*; Taura, Futoshi*; Arai, Shigeki; Blaber, M.*; Shoyama, Yukihiro*; Morimoto, Satoshi*; Kuroki, Ryota
Journal of Molecular Biology, 423(1), p.96 - 105, 2012/10
Times Cited Count:90 Percentile:89.94(Biochemistry & Molecular Biology)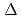 1-tetrahydrocannabinolic acid (THCA) synthase catalyzes the oxidative cyclization of cannabigerolic acid (CBGA) into THCA, the precursor of the primary psychoactive agent
1-tetrahydrocannabinolic acid (THCA) synthase catalyzes the oxidative cyclization of cannabigerolic acid (CBGA) into THCA, the precursor of the primary psychoactive agent 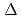 1-tetrahydrocannabinol in
1-tetrahydrocannabinol in 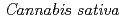 . The structure-function relationship of THCA synthase was investigated by X-ray structure determination (2.75
. The structure-function relationship of THCA synthase was investigated by X-ray structure determination (2.75  resolution) and mutational analysis. Specific amino acid residues were identified in the active site of THCA synthase that are involved in the oxidative cyclization of the CBGA substrate.
resolution) and mutational analysis. Specific amino acid residues were identified in the active site of THCA synthase that are involved in the oxidative cyclization of the CBGA substrate.
Ishibashi, Matsujiro*; Uchino, Manami*; Arai, Shigeki; Kuroki, Ryota; Arakawa, Tsutomu*; Tokunaga, Masao*
Archives of Biochemistry and Biophysics, 525(1), p.47 - 52, 2012/09
Times Cited Count:7 Percentile:19.80(Biochemistry & Molecular Biology)Nucleoside diphosphate kinase (HsNDK) from Halobacterium salinarum requires salt at high concentrations for folding. A D148C mutant, in which Asp148 was replaced with Cys, was designed to enhance stability and folding in low salt solution by S-S bond. It showed increased thermal stability by about 10 C in 0.2 M NaCl over the wild type HsNDK. It refolded from heat-denaturation even in 0.1 M NaCl, while the wild type required 2 M NaCl to achieve the same level of activity recovery. This enhanced refolding is due to the three S-S bonds between two basic dimeric units in the hexameric HsNDK structure. Moreover, salt concentration dependency of heat-denaturation process and refolding process of the wild type and D148C mutant HsNDKs were investigated by the circular dichroism and native-PAGE analysis.
C in 0.2 M NaCl over the wild type HsNDK. It refolded from heat-denaturation even in 0.1 M NaCl, while the wild type required 2 M NaCl to achieve the same level of activity recovery. This enhanced refolding is due to the three S-S bonds between two basic dimeric units in the hexameric HsNDK structure. Moreover, salt concentration dependency of heat-denaturation process and refolding process of the wild type and D148C mutant HsNDKs were investigated by the circular dichroism and native-PAGE analysis.
 sp. nucleoside diphosphate kinase
sp. nucleoside diphosphate kinaseArai, Shigeki; Yonezawa, Yasushi; Okazaki, Nobuo; Matsumoto, Fumiko; Tamada, Taro; Tokunaga, Hiroko*; Ishibashi, Matsujiro*; Blaber, M.; Tokunaga, Masao*; Kuroki, Ryota
Protein Science, 21(4), p.498 - 510, 2012/04
Times Cited Count:15 Percentile:34.81(Biochemistry & Molecular Biology)In order to clarify the oligomer state of nucleoside diphosphate kinase (NDK) from moderately halophilic  sp. 593 (HaNDK), the crystal structure of HaNDK was determined by X-ray crystallography. The crystal structures of the wild-type HaNDK and the mutant HaNDK (E134A) showed a dimer and a tetramer, respectively. The higher ordered association of proteins usually contributes to an increase in thermal stability and substrate affinity. The change in the assembly form by a minimum mutation may be an effective way for NDK to acquire molecular characteristics suited to various circumstances.
sp. 593 (HaNDK), the crystal structure of HaNDK was determined by X-ray crystallography. The crystal structures of the wild-type HaNDK and the mutant HaNDK (E134A) showed a dimer and a tetramer, respectively. The higher ordered association of proteins usually contributes to an increase in thermal stability and substrate affinity. The change in the assembly form by a minimum mutation may be an effective way for NDK to acquire molecular characteristics suited to various circumstances.
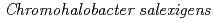 DSM3043; Both residues 134 and 136 are critical for the tetramer assembly
DSM3043; Both residues 134 and 136 are critical for the tetramer assemblyTokunaga, Hiroko*; Izutsu, Kenichi*; Arai, Shigeki; Yonezawa, Yasushi; Kuroki, Ryota; Arakawa, Tsutomu*; Tokunaga, Masao*
Enzyme and Microbial Technology, 46(2), p.129 - 135, 2010/02
Times Cited Count:6 Percentile:19.99(Biotechnology & Applied Microbiology)Both wild-type nucleoside diphosphate kinase from moderately halophilc 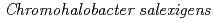 (CsNDK (GNE), GNE represents Gly134-Asn135-Glu136) and mutant CsNDK (ANE), both of which have a neutral amino acid at residue 134, were found to form a dimer. These constructs contain Glu136, which may also cause steric barrier and charge repulsion. A double mutant, CsNDK (ANT), having Thr at 136 resulted in stable tetrameric assembly, supporting the above notion. A mutant CsNDK (GNT) reverted, however, to a dimer again, indicating that the introduced Ala residue at 134th in the double mutant generated a hydrophobic cluster consisting of the Ala residues and thereby stabilized dimer-dimer association of CsNDK assembly, while Gly destabilized it due to the loss of this cluster. Based on these observations, it is evident that both residues 134 and 136 contribute to the subunit assembly of CsNDK.
(CsNDK (GNE), GNE represents Gly134-Asn135-Glu136) and mutant CsNDK (ANE), both of which have a neutral amino acid at residue 134, were found to form a dimer. These constructs contain Glu136, which may also cause steric barrier and charge repulsion. A double mutant, CsNDK (ANT), having Thr at 136 resulted in stable tetrameric assembly, supporting the above notion. A mutant CsNDK (GNT) reverted, however, to a dimer again, indicating that the introduced Ala residue at 134th in the double mutant generated a hydrophobic cluster consisting of the Ala residues and thereby stabilized dimer-dimer association of CsNDK assembly, while Gly destabilized it due to the loss of this cluster. Based on these observations, it is evident that both residues 134 and 136 contribute to the subunit assembly of CsNDK.
Adachi, Motoyasu; Ohara, Takashi; Kurihara, Kazuo; Tamada, Taro; Honjo, Eijiro; Okazaki, Nobuo; Arai, Shigeki; Shoyama, Yoshinari; Kimura, Kaname*; Matsumura, Hiroyoshi*; et al.
Proceedings of the National Academy of Sciences of the United States of America, 106(12), p.4641 - 4646, 2009/03
Times Cited Count:114 Percentile:90.42(Multidisciplinary Sciences)To further understand the catalytic mechanism and inhibitor recognition of HIV-1 protease, we need to determine the locations of key hydrogen atoms in the catalytic aspartates Asp25 and Asp125. The structure of HIV-1 protease in complex with transition-state analog KNI-272 was determined by combined neutron crystallography at 1.9  resolution and X-ray crystallography at 1.4
resolution and X-ray crystallography at 1.4  resolution. The resulting structural data shows that the catalytic residue Asp25 is protonated and that Asp125 is deprotonated. The proton on Asp25 makes a hydrogen bond with the carbonyl group of the allophenylnorstatine group in KNI-272. The deprotonated Asp125 bonds to the hydroxyl proton of Apns. The results provide direct experimental evidence for proposed aspects of the catalytic mechanism of HIV-1 protease; and can therefore contribute substantially to the development of specific inhibitors for therapeutic application.
resolution. The resulting structural data shows that the catalytic residue Asp25 is protonated and that Asp125 is deprotonated. The proton on Asp25 makes a hydrogen bond with the carbonyl group of the allophenylnorstatine group in KNI-272. The deprotonated Asp125 bonds to the hydroxyl proton of Apns. The results provide direct experimental evidence for proposed aspects of the catalytic mechanism of HIV-1 protease; and can therefore contribute substantially to the development of specific inhibitors for therapeutic application.
Tokunaga, Hiroko*; Ishibashi, Matsujiro*; Arisaka, Fimio*; Arai, Shigeki; Kuroki, Ryota; Arakawa, Tsutomu*; Tokunaga, Masao*
FEBS Letters, 582(7), p.1049 - 1054, 2008/04
Times Cited Count:17 Percentile:39.33(Biochemistry & Molecular Biology) nucleoside diphosphate kinase (HaNDK) forms a dimeric assembly and
nucleoside diphosphate kinase (HaNDK) forms a dimeric assembly and 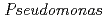 NDK (PaNDK) forms a tetrameric assembly. The mutation of Glu134 to Ala in HaNDK resulted in conversion of the native dimeric structure to the tetramer assembly. Conversely, the mutation of Ala134 to Glu in PaNDK leads to conversion from tetramer to dimer assembly, indicating that a single amino acid substitution at position 134 results in an alteration of the oligomeric structure of NDK. Modeling structure of HaNDK and PaNDK, based on the crystal structure of
NDK (PaNDK) forms a tetrameric assembly. The mutation of Glu134 to Ala in HaNDK resulted in conversion of the native dimeric structure to the tetramer assembly. Conversely, the mutation of Ala134 to Glu in PaNDK leads to conversion from tetramer to dimer assembly, indicating that a single amino acid substitution at position 134 results in an alteration of the oligomeric structure of NDK. Modeling structure of HaNDK and PaNDK, based on the crystal structure of 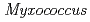 NDK, suggested sufficient repulsion by Glu134 to disrupt dimer-dimer interaction to form tetramer.
NDK, suggested sufficient repulsion by Glu134 to disrupt dimer-dimer interaction to form tetramer.
Arai, Shigeki; Yonezawa, Yasushi; Okazaki, Nobuo; Tamada, Taro; Tokunaga, Hiroko*; Ishibashi, Matsujiro*; Tokunaga, Masao*; Kuroki, Ryota
no journal, ,
The nucleoside diphosphate kinases (NDKs) are known to have a tetrameric or hexameric oligomer structure formed by association of common dimeric components. We determined the crystal structure of E134A mutant NDK from  sp. 593 (HaNDK) and found that two kinds of tetrameric assemblies, Type I seen in the
sp. 593 (HaNDK) and found that two kinds of tetrameric assemblies, Type I seen in the 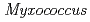 NDK tetramer and Type II seen in the
NDK tetramer and Type II seen in the 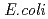 NDK tetramer, appeared in the asymmetric unit. Change in the assembly form may be an effective way for NDK to acquire molecular characteristics suited to various circumstances.
NDK tetramer, appeared in the asymmetric unit. Change in the assembly form may be an effective way for NDK to acquire molecular characteristics suited to various circumstances.
Adachi, Motoyasu; Ohara, Takashi; Kurihara, Kazuo; Tamada, Taro; Honjo, Eijiro; Okazaki, Nobuo; Arai, Shigeki; Shoyama, Yoshinari; Kimura, Kaname*; Matsumura, Hiroyoshi*; et al.
no journal, ,
HIV-1 protease is a dimeric aspartic protease that cleaves the nascent polyproteins of HIV-1 and plays an essential role in viral replication. To further understand the catalytic mechanism of HIV-1 protease, we have determined the crystal structure of HIV-1 protease in complex with a transition state mimetic tripeptide inhibitor, KNI-272 to 1.9  resolution by neutron crystallography in combination with 1.4
resolution by neutron crystallography in combination with 1.4  resolution X-ray diffraction data. Our results indicates that the carbonyl group of allophenylnorstatine in KNI-272 forms a significant hydrogen bond with protonated Asp 25, and the hydrogen atom from the hydroxyl group of Apns forms a remarkable hydrogen bond with the deprotonated Asp125. These results show direct evidence that Asp25 provides a proton to carbonyl group of substrate and Asp125 contributes to activate the attacking water molecule as a nucleophile.
resolution X-ray diffraction data. Our results indicates that the carbonyl group of allophenylnorstatine in KNI-272 forms a significant hydrogen bond with protonated Asp 25, and the hydrogen atom from the hydroxyl group of Apns forms a remarkable hydrogen bond with the deprotonated Asp125. These results show direct evidence that Asp25 provides a proton to carbonyl group of substrate and Asp125 contributes to activate the attacking water molecule as a nucleophile.
Tamada, Taro; Arai, Shigeki; Honjo, Eijiro; Kuroki, Ryota
no journal, ,
no abstracts in English
Tamada, Taro; Honjo, Eijiro; Arai, Shigeki; Kuroki, Ryota
no journal, ,
The ligand-receptor interaction in the extracellular environment is essential for signal transduction of biological processes. We have focused on the structure/function relationships of these proteins. Granulocyte colony-stimulating factor (GCSF) has become an important cytokine for medical treatment of patients suffering from granulopoenia through regulating the maturation, proliferation, and differentiation of the precursor cells of neutrophilic granulocytes. We determined a crystal structure of the signaling complex between human GCSF and a ligand binding region of GCSF receptor (GCSF-R). We also determined additional structures of human cytokines and extracellular regions of cytokine receptors, where the complexation with an antibody fragment (Fab) was used for crystallization and structure determination.
Adachi, Motoyasu; Ohara, Takashi; Kurihara, Kazuo; Tamada, Taro; Honjo, Eijiro*; Okazaki, Nobuo; Arai, Shigeki; Shoyama, Yoshinari*; Matsumura, Hiroyoshi*; Adachi, Hiroaki*; et al.
no journal, ,
To understand the catalytic mechanism of HIV-1 protease, we have determined the crystal structure of HIV-1 protease in complex with a transition state mimetic inhibitor, KNI-272 by neutron crystallography. Our results indicates that the carbonyl group of allophenylnorstatine in KNI-272 forms a significant hydrogen bond with protonated Asp 25, and the hydrogen atom from the hydroxyl group of allophenylnorstatine forms a remarkable hydrogen bond with the deprotonated Asp125. These results show direct evidence that Asp25 provides a proton to carbonyl group of substrate and Asp125 contributes to activate the attacking water molecule as a nucleophile.
Arai, Shigeki; Tamada, Taro; Maeda, Yoshitake*; Kuroki, Ryota
no journal, ,
The mouse antibody TN1 recognizes the human thrombopoietin (hTPO) that primarily stimulates megakaryocytopoiesis and platelet production. In order to clarify the mechanism of the neutralizing activity of the TN1 antibody, the crystal structure of TN1-Fab was determined to 2.0 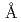 resolution and was compared with that of TN1-Fab / hTPO complex (PDB id 1V7M and 1V7N). The relative angle of the variable- and constant-regions of the hTPO unbound form of TN1-Fab was slightly shifted from those of TN1-Fab / hTPO complex (rms deviation = 2.4
resolution and was compared with that of TN1-Fab / hTPO complex (PDB id 1V7M and 1V7N). The relative angle of the variable- and constant-regions of the hTPO unbound form of TN1-Fab was slightly shifted from those of TN1-Fab / hTPO complex (rms deviation = 2.4 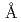 for all C
for all C atoms of Fab). On the other hand, only the slight shift of the side chain of CDR was observed (rmsd
atoms of Fab). On the other hand, only the slight shift of the side chain of CDR was observed (rmsd 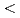 1.5
1.5 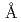 for atoms of side chain of each CDR) upon recognition of the epitope of hTPO. This structural shift of side chain of paratope did not accompany the
for atoms of side chain of each CDR) upon recognition of the epitope of hTPO. This structural shift of side chain of paratope did not accompany the 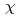 -angle rotation, but the parallel shift. These results indicate that the CDR of TN1-Fab need not accompany the large conformational changes of upon TPO recognition.
-angle rotation, but the parallel shift. These results indicate that the CDR of TN1-Fab need not accompany the large conformational changes of upon TPO recognition.