Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Tashiro, Koji*; Kusaka, Katsuhiro*; Yamamoto, Hiroko*; Hosoya, Takaaki*; Okada, Shuji*; Ohara, Takashi
Polymers (Internet), 15(2), p.465_1 - 465_44, 2023/01
Times Cited Count:3 Percentile:11.04(Polymer Science)Yano, Naomine*; Yamada, Taro*; Hosoya, Takaaki*; Ohara, Takashi; Tanaka, Ichiro*; Niimura, Nobuo*; Kusaka, Katsuhiro*
Acta Crystallographica Section D; Structural Biology (Internet), 74(11), p.1041 - 1052, 2018/11
Times Cited Count:15 Percentile:74.07(Biochemical Research Methods)Tashiro, Koji*; Kusaka, Katsuhiro*; Hosoya, Takaaki*; Ohara, Takashi; Hanesaka, Makoto*; Yoshizawa, Yoshinori*; Yamamoto, Hiroko*; Niimura, Nobuo*; Tanaka, Ichiro*; Kurihara, Kazuo*; et al.
Macromolecules, 51(11), p.3911 - 3922, 2018/06
Times Cited Count:6 Percentile:18.61(Polymer Science)Nakajima, Kenji; Kawakita, Yukinobu; Ito, Shinichi*; Abe, Jun*; Aizawa, Kazuya; Aoki, Hiroyuki; Endo, Hitoshi*; Fujita, Masaki*; Funakoshi, Kenichi*; Gong, W.*; et al.
Quantum Beam Science (Internet), 1(3), p.9_1 - 9_59, 2017/12
The neutron instruments suite, installed at the spallation neutron source of the Materials and Life Science Experimental Facility (MLF) at the Japan Proton Accelerator Research Complex (J-PARC), is reviewed. MLF has 23 neutron beam ports and 21 instruments are in operation for user programs or are under commissioning. A unique and challenging instrumental suite in MLF has been realized via combination of a high-performance neutron source, optimized for neutron scattering, and unique instruments using cutting-edge technologies. All instruments are/will serve in world-leading investigations in a broad range of fields, from fundamental physics to industrial applications. In this review, overviews, characteristic features, and typical applications of the individual instruments are mentioned.
Yano, Naomine*; Yamada, Taro*; Hosoya, Takaaki*; Ohara, Takashi; Tanaka, Ichiro*; Kusaka, Katsuhiro*
Scientific Reports (Internet), 6, p.36628_1 - 36628_9, 2016/12
Times Cited Count:12 Percentile:60.96(Multidisciplinary Sciences)Unno, Masayoshi*; Sugishima, Masakazu*; Wada, Kei*; Hagiwara, Yoshinori*; Kusaka, Katsuhiro*; Tamada, Taro; Fukuyama, Keiichi*
Nihon Kessho Gakkai-Shi, 57(5), p.297 - 303, 2015/10
Bilin compounds are fundamentally important for oxygenic photosynthetic organisms, because they are utilized as pigments for photosynthesis (phycobilins) and photoreceptors (phytochromobilin). Phycocyanobilin (PCB), a phycobilin, comprises the chromophore of algal phytochromes and the core phycobiliprotein antennae of cyanobacteria and red algae. PCB is biosynthesized by a member of the ferredoxin-dependent bilin reductase family, phycocyanobilin:ferredoxin oxidoreductase (PcyA). In the present study, we determined the neutron crystal structure of PcyA in complex with its substrate biliverdin (BV). This neutron structure revealed the protonation state of BV and the surrounding residues. We found that two forms of BV, neutral BV and protonated BVH , were coupled with the two conformation/protonation states of the essential residue Asp105. Further, His88 and His74 near BV were singly protonated and were connected with an intervening hydronium ion. Neutron analysis also revealed how X-ray irradiation of the PcyA-BV crystal altered the structure of the PcyA-BV complex.
, were coupled with the two conformation/protonation states of the essential residue Asp105. Further, His88 and His74 near BV were singly protonated and were connected with an intervening hydronium ion. Neutron analysis also revealed how X-ray irradiation of the PcyA-BV crystal altered the structure of the PcyA-BV complex.
Unno, Masayoshi*; Ishikawa, Kumiko*; Kusaka, Katsuhiro*; Tamada, Taro; Hagiwara, Yoshinori*; Sugishima, Masakazu*; Wada, Kei*; Yamada, Taro*; Tomoyori, Katsuaki; Hosoya, Takaaki*; et al.
Journal of the American Chemical Society, 137(16), p.5452 - 5460, 2015/04
Times Cited Count:29 Percentile:62.81(Chemistry, Multidisciplinary)Phycocyanobilin, a light-harvesting and photoreceptor pigment in higher plants, algae, and cyanobacteria, is synthesized from biliverdin IX (BV) by phycocyanobilin:ferredoxin oxidoreductase (PcyA) via two steps of two-proton-coupled two-electron reduction. We determined the neutron structure of PcyA from cyanobacteria complexed with BV, revealing the exact location of the hydrogen atoms involved in catalysis. Notably, approximately half of the BV bound to PcyA was BVH
(BV) by phycocyanobilin:ferredoxin oxidoreductase (PcyA) via two steps of two-proton-coupled two-electron reduction. We determined the neutron structure of PcyA from cyanobacteria complexed with BV, revealing the exact location of the hydrogen atoms involved in catalysis. Notably, approximately half of the BV bound to PcyA was BVH , a state in which all four pyrrole nitrogen atoms were protonated. The protonation states of BV complemented the protonation of adjacent Asp105. The "axial "water molecule that interacts with the neutral pyrrole nitrogen of the A-ring was identified. His88 N
, a state in which all four pyrrole nitrogen atoms were protonated. The protonation states of BV complemented the protonation of adjacent Asp105. The "axial "water molecule that interacts with the neutral pyrrole nitrogen of the A-ring was identified. His88 N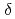 was protonated to form a hydrogen bond with the lactam O atom of the BV A-ring. His88 and His74 were linked by hydrogen bonds via H
was protonated to form a hydrogen bond with the lactam O atom of the BV A-ring. His88 and His74 were linked by hydrogen bonds via H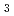 O
O . These results imply that Asp105, His88, and the axial water molecule contribute to proton transfer during PcyA catalysis.
. These results imply that Asp105, His88, and the axial water molecule contribute to proton transfer during PcyA catalysis.
Kusaka, Katsuhiro*; Ohara, Takashi; Yamada, Taro*; Kiyanagi, Ryoji
Hamon, 25(2), p.171 - 178, 2015/00
Tashiro, Koji*; Hanesaka, Makoto*; Yamamoto, Hiroko*; Wasanasuk, K.*; Jayaratri, P.*; Yoshizawa, Yoshinori*; Tanaka, Ichiro*; Niimura, Nobuo*; Kusaka, Katsuhiro*; Hosoya, Takaaki*; et al.
Kobunshi Rombunshu, 71(11), p.508 - 526, 2014/11
Times Cited Count:6 Percentile:20.64(Polymer Science)The crystal structure analysis of various polymer substances has been reviewed on the basis of wide-angle high-energy X-ray and neutron diffraction data. The progress in structural analytical techniques of polymer crystals have been reviewed at first. The structural models proposed so far were reinvestigated and new models have been proposed for various kinds of polymer crystals including polyethylene, poly(vinyl alcohol), poly(lactic acid) and its stereocomplex etc. The hydrogen atomic positions were also clarified by the quantitative analysis of wide-angle neutron diffraction data, from which the physical properties of polymer crystals have been evaluated theoretically. The bonded electron density distribution has been estimated for a polydiacetylene single crystal on the basis of the so-called X-N method or by the combination of structural information derived from X-ray and neutron diffraction data analysis. Some comments have been added about future developments in the field of structure-property relationship determination.
Tomoyori, Katsuaki; Kusaka, Katsuhiro*; Yamada, Taro*; Tamada, Taro
Journal of Structural and Functional Genomics, 15(3), p.131 - 135, 2014/09
We plan to design a high-resolution biomacromolecule neutron time-of-flight diffractometer, which allows us to collect data from crystals with unit cells above 250  in the Materials and Life Science Experimental Facility at the Japan Proton Accelerator Research Complex. This new diffractometer can be used for a detailed analysis of large proteins such as membrane proteins and supermolecular complex. A quantitative comparison of the intensity and pulse width of a decoupled moderator (DM) against a coupled moderator (CM) considering the pulse width time resolution indicated that the DM satisfies the criteria for our diffractometer rather than the CM. The results suggested that a characteristic feature of the DM, i.e., narrow pulse width with a short tail, is crucial for the separation of Bragg reflections from crystals with large unit cells. On the other hand, it should be noted that the weak signals from the DM are buried under the high-level background caused by the incoherent scattering of hydrogen atoms, especially, in the case of large unit cells. We propose a profile-fitting integration method combined with the energy loss functions and a background subtraction method achieved by employing the Statistics-sensitive Nonlinear Iterative Peak-clipping algorithm.
in the Materials and Life Science Experimental Facility at the Japan Proton Accelerator Research Complex. This new diffractometer can be used for a detailed analysis of large proteins such as membrane proteins and supermolecular complex. A quantitative comparison of the intensity and pulse width of a decoupled moderator (DM) against a coupled moderator (CM) considering the pulse width time resolution indicated that the DM satisfies the criteria for our diffractometer rather than the CM. The results suggested that a characteristic feature of the DM, i.e., narrow pulse width with a short tail, is crucial for the separation of Bragg reflections from crystals with large unit cells. On the other hand, it should be noted that the weak signals from the DM are buried under the high-level background caused by the incoherent scattering of hydrogen atoms, especially, in the case of large unit cells. We propose a profile-fitting integration method combined with the energy loss functions and a background subtraction method achieved by employing the Statistics-sensitive Nonlinear Iterative Peak-clipping algorithm.
Kusaka, Katsuhiro*; Hosoya, Takaaki*; Yamada, Taro*; Tomoyori, Katsuaki; Ohara, Takashi; Katagiri, Masaki*; Kurihara, Kazuo; Tanaka, Ichiro*; Niimura, Nobuo*
Journal of Synchrotron Radiation, 20(6), p.994 - 998, 2013/11
Times Cited Count:39 Percentile:85.62(Instruments & Instrumentation)The IBARAKI biological crystal diffractometer, iBIX, is a high-performance time-of-flight neutron single-crystal diffractometer for elucidating mainly the hydrogen, protonation and hydration structures of biological macromolecules in various life processes. Since the end of 2008, iBIX has been available to user's experiments supported by Ibaraki University. Since August 2012, an upgrade of the 14-existing detectors has begun and 16 new detectors have been installed for iBIX. The total measurement efficiency of the present diffractometer has been impoved by one order of magnitude from the previous one with the increasing of accelerator power. In December 2012, commissioning of the new detectors was successful, and collection of the diffraction dataset of ribonucrease A as a standard protein was attempted in order to estimate the performance of the upgraded iBIX in comparison with previous results. The resolution of diffraction data, equivalence among intensities of symmetry-related reflections and reliability of the refined structure have been improved dramatically. iBIX is expected to be one of the highest-performance neutron single-crystal diffractometers for biological macromolecules in the world.
Yokoyama, Takeshi*; Mizuguchi, Mineyuki*; Nabeshima, Yuko*; Kusaka, Katsuhiro*; Yamada, Taro*; Hosoya, Takaaki*; Ohara, Takashi; Kurihara, Kazuo; Tanaka, Ichiro*; Niimura, Nobuo*
Journal of Synchrotron Radiation, 20(6), p.834 - 837, 2013/11
Times Cited Count:7 Percentile:36.21(Instruments & Instrumentation)Transthyretin (TTR) is a tetrameric protein. TTR misfolding and aggregation are associated with human amyloid diseases. Dissociation of the TTR tetramer is believed to be the rate-limiting step in the amyloid fibril formation cascade. Low pH is known to promote dissociation into monomer and the formation of amyloid fibrils. In order to reveal the molecular mechanisms underlying pH sensitivity and structural stabilities of TTR, neutron diffraction studies were conducted using the IBARAKI Biological Crystal Diffractometer with the time-of-flight method. Crystals for the neutron diffraction experiments were grown up to 2.5 mm for four months. The neutron crystal structure solved at 2.0
for four months. The neutron crystal structure solved at 2.0  revealed the protonation states of His88 and the detailed hydrogen-bond network depending on the protonation states of His88. This hydrogen-bond network is involved in monomer-monomer and dimer-dimer interactions, suggesting that the double protonation of His88 by acidification breaks the hydrogen-bond network and causes the destabilization of the TTR tetramer. Structural comparison with the X-ray crystal structure at acidic pH identified the three amino acid residues responsible for the pH sensitivity of TTR. Our neutron model provides insights into the molecular stability related to amyloidosis.
revealed the protonation states of His88 and the detailed hydrogen-bond network depending on the protonation states of His88. This hydrogen-bond network is involved in monomer-monomer and dimer-dimer interactions, suggesting that the double protonation of His88 by acidification breaks the hydrogen-bond network and causes the destabilization of the TTR tetramer. Structural comparison with the X-ray crystal structure at acidic pH identified the three amino acid residues responsible for the pH sensitivity of TTR. Our neutron model provides insights into the molecular stability related to amyloidosis.
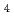 H
H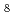 ONH
ONH PbBr
PbBr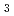 studied using single-crystal neutron diffraction
studied using single-crystal neutron diffractionKawasaki, Takuro; Takahashi, Miwako*; Ohara, Takashi*; Tanaka, Ichiro*; Kusaka, Katsuhiro*; Hosoya, Takaaki*; Yamada, Taro*; Kurihara, Kazuo
Journal of the Physical Society of Japan, 81(9), p.094602_1 - 094602_6, 2012/09
Times Cited Count:7 Percentile:45.48(Physics, Multidisciplinary)Yokoyama, Takeshi*; Mizuguchi, Mineyuki*; Nabeshima, Yuko*; Kusaka, Katsuhiro*; Yamada, Taro*; Hosoya, Takaaki*; Ohara, Takashi*; Kurihara, Kazuo; Tomoyori, Katsuaki*; Tanaka, Ichiro*; et al.
Journal of Structural Biology, 177(2), p.283 - 290, 2012/02
Times Cited Count:49 Percentile:82.40(Biochemistry & Molecular Biology)Tanaka, Ichiro*; Kusaka, Katsuhiro*; Hosoya, Takaaki*; Niimura, Nobuo*; Ohara, Takashi*; Kurihara, Kazuo; Yamada, Taro*; Onishi, Yuki*; Tomoyori, Katsuaki*; Yokoyama, Takeshi*
Acta Crystallographica Section D, 66(11), p.1194 - 1197, 2010/11
Times Cited Count:52 Percentile:94.69(Biochemical Research Methods)The IBARAKI Biological Crystal Diffractometer (iBIX), a new diffractometer for protein crystallography at the next-generation neutron source at J-PARC (Japan Proton Accelerator Research Complex), has been constructed and has been operational since December 2008. Preliminary structure analyses of organic crystals showed that iBIX has high performance even at 120 kW operation and the first full data set is being collected from a protein crystal.
Tanaka, Ichiro*; Kusaka, Katsuhiro*; Hosoya, Takaaki*; Ohara, Takashi*; Kurihara, Kazuo; Niimura, Nobuo*
Yakugaku Zasshi, 130(5), p.665 - 670, 2010/05
Times Cited Count:0 Percentile:0.00(Pharmacology & Pharmacy)Ibaraki Prefectural Government together with Ibaraki University and Japan Atomic Energy Agency (JAEA) has almost finished constructing a time-of-flight (TOF) neutron diffractometer for biological macromolecules for industrial use at J-PARC, IBARAKI Biological Crystal Diffractometer (iBIX). Since 2009, Ibaraki University has been asked to operate this machine in order for users to do experiments by Ibaraki Prefecture. The diffractometer is designed to cover sample crystals which have their cell edges up to around 150  . It is expected to measure more than 100 samples per year if they have 2 mm
. It is expected to measure more than 100 samples per year if they have 2 mm in crystal volume, and to measure even around 0.1 mm
in crystal volume, and to measure even around 0.1 mm in crystal volume of biological samples. The efficiency of iBIX is also expected about 100 times larger than those of the present high performance diffractometers at JRR-3 in JAEA when 1MW power realizes in J-PARC. Since December 2008, iBIX has been open to users and several proteins and organic compounds were tested under 20 kW proton power of J-PARC. It was found that one of their proteins was diffracted up to 1.4
in crystal volume of biological samples. The efficiency of iBIX is also expected about 100 times larger than those of the present high performance diffractometers at JRR-3 in JAEA when 1MW power realizes in J-PARC. Since December 2008, iBIX has been open to users and several proteins and organic compounds were tested under 20 kW proton power of J-PARC. It was found that one of their proteins was diffracted up to 1.4  in d-spacing, which was nearly comparable resolution to that of BIX-3 in JRR-3 when used the same crystal as at iBIX for reasonable exposure time. In May 2009, 14 detector units were set up. By the end of fiscal year 2009, the basic part of data reduction software will be finished and an equipment blowing low temperature gas to the sample will be installed with the cooperation of JAEA.
in d-spacing, which was nearly comparable resolution to that of BIX-3 in JRR-3 when used the same crystal as at iBIX for reasonable exposure time. In May 2009, 14 detector units were set up. By the end of fiscal year 2009, the basic part of data reduction software will be finished and an equipment blowing low temperature gas to the sample will be installed with the cooperation of JAEA.
Ohara, Takashi; Kurihara, Kazuo; Nakatani, Takeshi; Suzuki, Jiro*; Otomo, Toshiya*; Hosoya, Takaaki*; Kusaka, Katsuhiro*; Tanaka, Ichiro*; Niimura, Nobuo*; Seki, Akiyuki; et al.
no journal, ,
no abstracts in English
Ohara, Takashi; Kusaka, Katsuhiro*; Hosoya, Takaaki*; Kurihara, Kazuo; Tomoyori, Katsuaki*; Niimura, Nobuo*; Tanaka, Ichiro*; Suzuki, Jiro*; Nakatani, Takeshi; Otomo, Toshiya*; et al.
no journal, ,
no abstracts in English
Ohara, Takashi; Kusaka, Katsuhiro*; Hosoya, Takaaki; Kurihara, Kazuo; Tomoyori, Katsuaki*; Niimura, Nobuo*; Tanaka, Ichiro*; Suzuki, Jiro*; Nakatani, Takeshi; Otomo, Toshiya*; et al.
no journal, ,
For a single crystal diffractometer, a data reduction software which extracts a HKLF list from the raw data is one of the most important components. We are now developing a new data reduction software for a new single crystal neutron diffractometer, "IBARAKI Biological Crystal Diffractometer (iBIX)", which is now constructing at the MLF of J-PARC.
Ohara, Takashi; Kurihara, Kazuo; Kusaka, Katsuhiro; Hosoya, Takaaki; Tanaka, Ichiro*; Niimura, Nobuo*; Ozeki, Tomoji*; Aizawa, Kazuya; Morii, Yukio; Arai, Masatoshi; et al.
no journal, ,
Ibaraki Prefectural Government in Japan has started to construct a TOF single crystal neutron diffractometer for biological macromolecules for industrial use at J-PARC. For this diffractometer, design of an efficient neutron transportation system is important because this diffractometer has 40m source-sample distance. Recently, we designed a supermirror neutron guide which can transport 0.7-3.8 Angstrom neutron efficiently. The total length of the mirror section is 25m. At the first 17m, the mirror has curvature (R=4300m) for horizontal direction in order to remove high-energy neutron and  ray. Simultaneously, all of the mirror section has tapered angle for vertical direction in order to reduce the frequency of neutron reflection at the surface of the supermirror. The neutron flux and profile at the sample position was calculated by Monte Carlo simulation softwares, McStas and IDEAS and compared with a curved, non-tapered guide we had designed previously. In result, the new supermirror system has 2 times gain for 0.7 Angstrom neutron and 1.5 times for 1.5 Angstrom neutron, and the beam profile at the sample position has a rectangular shape.
ray. Simultaneously, all of the mirror section has tapered angle for vertical direction in order to reduce the frequency of neutron reflection at the surface of the supermirror. The neutron flux and profile at the sample position was calculated by Monte Carlo simulation softwares, McStas and IDEAS and compared with a curved, non-tapered guide we had designed previously. In result, the new supermirror system has 2 times gain for 0.7 Angstrom neutron and 1.5 times for 1.5 Angstrom neutron, and the beam profile at the sample position has a rectangular shape.