Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
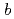
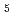 reductase by X-ray crystallography; New insights into regulation of efficient electron transfer
reductase by X-ray crystallography; New insights into regulation of efficient electron transferYamada, Mitsugu*; Tamada, Taro; Takeda, Kazuki*; Matsumoto, Fumiko*; Ono, Hiraku*; Kosugi, Masayuki*; Takaba, Kiyofumi*; Shoyama, Yoshinari*; Kimura, Shigenobu*; Kuroki, Ryota; et al.
Journal of Molecular Biology, 425(22), p.4295 - 4306, 2013/11
Times Cited Count:24 Percentile:52.94(Biochemistry & Molecular Biology)NADH-Cytochrome 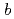
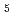 reductase (b5R), a flavoprotein consisting of NADH and flavin adenine dinucleotide (FAD) binding domains, catalyzes electron transfer from the two-electron carrier NADH to the one-electron carrier cytochrome
reductase (b5R), a flavoprotein consisting of NADH and flavin adenine dinucleotide (FAD) binding domains, catalyzes electron transfer from the two-electron carrier NADH to the one-electron carrier cytochrome 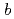
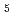 (Cb5). The crystal structures of both the fully reduced form and the oxidized form of porcine liver b5R were determined. In the reduced b5R structure determined at 1.68
(Cb5). The crystal structures of both the fully reduced form and the oxidized form of porcine liver b5R were determined. In the reduced b5R structure determined at 1.68 resolution, the relative configuration of the two domains was slightly shifted in comparison with that of the oxidized form. This shift resulted in an increase in the solvent-accessible surface area of FAD and created a new hydrogen-bonding interaction between the N5 atom of the isoalloxazine ring of FAD and the hydroxyl oxygen atom of Thr66, which is considered to be a key residue in the release of a proton from the N5 atom. The isoalloxazine ring of FAD in the reduced form is flat as in the oxidized form and stacked together with the nicotinamide ring of NAD
resolution, the relative configuration of the two domains was slightly shifted in comparison with that of the oxidized form. This shift resulted in an increase in the solvent-accessible surface area of FAD and created a new hydrogen-bonding interaction between the N5 atom of the isoalloxazine ring of FAD and the hydroxyl oxygen atom of Thr66, which is considered to be a key residue in the release of a proton from the N5 atom. The isoalloxazine ring of FAD in the reduced form is flat as in the oxidized form and stacked together with the nicotinamide ring of NAD . Determination of the oxidized b5R structure, including the hydrogen atoms, determined at 0.78
. Determination of the oxidized b5R structure, including the hydrogen atoms, determined at 0.78 resolution revealed the details of a hydrogen-bonding network from the N5 atom of FAD to His49 via Thr66. Both of the reduced and oxidized b5R structures explain how backflow in this catalytic cycle is prevented and the transfer of electrons to one-electron acceptors such as Cb5 is accelerated. Furthermore, crystallographic analysis by the cryo-trapping method suggests that re-oxidation follows a two-step mechanism. These results provide structural insights into the catalytic cycle of b5R.
resolution revealed the details of a hydrogen-bonding network from the N5 atom of FAD to His49 via Thr66. Both of the reduced and oxidized b5R structures explain how backflow in this catalytic cycle is prevented and the transfer of electrons to one-electron acceptors such as Cb5 is accelerated. Furthermore, crystallographic analysis by the cryo-trapping method suggests that re-oxidation follows a two-step mechanism. These results provide structural insights into the catalytic cycle of b5R.
 reductase at J-PARC
reductase at J-PARCHirano, Yu; Yamada, Mitsugu*; Kurihara, Kazuo; Kusaka, Katsuhiro*; Kimura, Shigenobu*; Miki, Kunio*; Tamada, Taro
no journal, ,
NADH-cytochrome  reductase (b5R), a flavoprotein consisting of NADH- and FAD-domains, catalyzes electron transfer from the two-electron carrier NADH to the one-electron carrier cytochrome
reductase (b5R), a flavoprotein consisting of NADH- and FAD-domains, catalyzes electron transfer from the two-electron carrier NADH to the one-electron carrier cytochrome  . The reaction catalyzed by b5R plays a role in fatty acid synthesis, cholesterol synthesis, and xenobiotic oxidation as a member of the electron transport chain on the endoplasmic reticulum. We have already determined the crystal structures of both the fully reduced and oxidized forms of porcine liver b5R by X-ray crystallography, but, its detail mechanism, especially hydride/proton transfers and exact states of semiquinone, still remains unknown. The hydrogen information obtained by neutron crystallography will be essential for the real understanding of catalytic cycle of the b5R. For neutron diffraction experiments, we prepared large crystals with the size of almost 2 mm
. The reaction catalyzed by b5R plays a role in fatty acid synthesis, cholesterol synthesis, and xenobiotic oxidation as a member of the electron transport chain on the endoplasmic reticulum. We have already determined the crystal structures of both the fully reduced and oxidized forms of porcine liver b5R by X-ray crystallography, but, its detail mechanism, especially hydride/proton transfers and exact states of semiquinone, still remains unknown. The hydrogen information obtained by neutron crystallography will be essential for the real understanding of catalytic cycle of the b5R. For neutron diffraction experiments, we prepared large crystals with the size of almost 2 mm . Large crystals were transferred to cryo-protectant solution by stepwise soaking method, and then were flash-frozen in a cold nitrogen gas stream. In preliminary neutron experiment, we confirmed some diffraction spots above 1.4
. Large crystals were transferred to cryo-protectant solution by stepwise soaking method, and then were flash-frozen in a cold nitrogen gas stream. In preliminary neutron experiment, we confirmed some diffraction spots above 1.4  , resolution from a crystal with the size of 1.8 mm
, resolution from a crystal with the size of 1.8 mm under 100 K at BL03 (iBIX), MLF, J-PARC after 14 hours exposure at 300 KW accelerator power.
under 100 K at BL03 (iBIX), MLF, J-PARC after 14 hours exposure at 300 KW accelerator power.
Hirano, Yu; Takeda, Kazuki*; Kurihara, Kazuo; Tamada, Taro; Miki, Kunio*
no journal, ,
It is important for understanding the electron transfer reaction to include the information about valence shell electrons and hydrogen atoms into crystal structure refinement. Recently, we have successfully collected 0.48 resolution data of high-potential iron-sulfur protein (HiPIP) in BL41XU beamline of SPring-8. We performed multipolar refinement with the
resolution data of high-potential iron-sulfur protein (HiPIP) in BL41XU beamline of SPring-8. We performed multipolar refinement with the 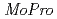 program to consider valence shell electrons in the structure refinement of HiPIP. After multipolar refinement, the deformation map clearly displays the distribution of valence shell electrons such as lone-pair electrons of carbonyl oxygen atoms, bonding electrons in aromatic rings, and
program to consider valence shell electrons in the structure refinement of HiPIP. After multipolar refinement, the deformation map clearly displays the distribution of valence shell electrons such as lone-pair electrons of carbonyl oxygen atoms, bonding electrons in aromatic rings, and 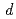 -orbital electrons of Fe atoms in the Fe
-orbital electrons of Fe atoms in the Fe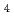 S
S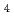 cluster. In addition, we performed preliminary neutron diffraction experiment at iBIX beamline of Japan Proton Accelerator Research Complex (J-PARC) and observed diffraction spots up to 1.17
cluster. In addition, we performed preliminary neutron diffraction experiment at iBIX beamline of Japan Proton Accelerator Research Complex (J-PARC) and observed diffraction spots up to 1.17 resolution using HiPIP crystal with the size of 2.3 mm
resolution using HiPIP crystal with the size of 2.3 mm .
.
 reductase
reductaseHirano, Yu; Yamada, Mitsugu*; Kurihara, Kazuo; Shoyama, Yoshinari*; Kuroki, Ryota; Kusaka, Katsuhiro*; Kimura, Shigenobu*; Takeda, Kazuki*; Miki, Kunio*; Tamada, Taro
no journal, ,
NADH-cytochrome  reductase (b5R), a flavoprotein consisting of NADH- and FAD- domains, catalyzes electron transfer from the two-electron carrier NADH to the one-electron carrier cytochrome
reductase (b5R), a flavoprotein consisting of NADH- and FAD- domains, catalyzes electron transfer from the two-electron carrier NADH to the one-electron carrier cytochrome  . The reaction catalyzed by
. The reaction catalyzed by  plays a role in fatty acid synthesis, cholesterol synthesis, and xenobiotic oxidation as a member of the electron transport chain on the endoplasmic reticulum. We have already determined the crystal structures of both the fully reduced and oxidized forms of porcine liver b5R by X-ray crystallography, but, its detail mechanism, especially hydride/proton transfers and exact states of semiquinone, still remains unknown. The hydrogen information obtained by neutron crystallography will be essential for the real understanding of catalytic cycle of the
plays a role in fatty acid synthesis, cholesterol synthesis, and xenobiotic oxidation as a member of the electron transport chain on the endoplasmic reticulum. We have already determined the crystal structures of both the fully reduced and oxidized forms of porcine liver b5R by X-ray crystallography, but, its detail mechanism, especially hydride/proton transfers and exact states of semiquinone, still remains unknown. The hydrogen information obtained by neutron crystallography will be essential for the real understanding of catalytic cycle of the  . A large crystal with the size of almost 2 mm
. A large crystal with the size of almost 2 mm was transferred to cryo-protectant solution by stepwise soaking method, and then were flash-frozen in a cold nitrogen gas stream. Using this crystal, we collected neutron data to 1.4
was transferred to cryo-protectant solution by stepwise soaking method, and then were flash-frozen in a cold nitrogen gas stream. Using this crystal, we collected neutron data to 1.4 resolution at BL03 (iBIX), MLF, J-PARC, and then collected to 0.85
resolution at BL03 (iBIX), MLF, J-PARC, and then collected to 0.85 resolution at BL5A, PF, KEK. Crystallographic refinement using both neutron and X-ray data is in progress.
resolution at BL5A, PF, KEK. Crystallographic refinement using both neutron and X-ray data is in progress.
Hirano, Yu; Tamada, Taro; Kurihara, Kazuo; Kusaka, Katsuhiro*; Miki, Kunio*
no journal, ,
High-potential iron-sulfur protein (HiPIP) possesses a 4Fe-4S cluster as a cofactor that shows 2+/3+ redox states. HiPIP is a soluble electron carrier protein that works in the photosynthetic electron transfer chain of purple photosynthetic bacteria. We have determined the neutron crystal structure of HiPIP at high resolution to obtain structure information including hydrogen atoms. Neutron diffraction experiment was performed at J-PARC, and the diffraction data set was obtained at 1.1  resolution that is the highest resolution in the protein neutron diffraction data. To perform X-ray and neutron joint refinement, we also collected X-ray diffraction data at Photon Factory, and the diffraction data set was obtained at 0.66
resolution that is the highest resolution in the protein neutron diffraction data. To perform X-ray and neutron joint refinement, we also collected X-ray diffraction data at Photon Factory, and the diffraction data set was obtained at 0.66  resolution.
resolution.
Hirano, Yu; Kurihara, Kazuo; Tamada, Taro; Kusaka, Katsuhiro*; Miki, Kunio*
no journal, ,
Many electron transfer proteins possess prosthetic groups, such as heme, flavin and iron-sulfur cluster. The electron transfer reaction is associated with the movement of hydrogen atoms or outer electrons of the prosthetic groups. We have determined high-resolution neutron structure of both high-potential iron-sulfur protein (HiPIP) and NADH cytochrome  reductase (b5R). We have collected neutron diffraction data of HiPIP and b5R at iBIX beamline of J-PARC, and diffraction data sets were obtained at 1.1
reductase (b5R). We have collected neutron diffraction data of HiPIP and b5R at iBIX beamline of J-PARC, and diffraction data sets were obtained at 1.1  resolution (HiPIP) and 1.4
resolution (HiPIP) and 1.4  resolution (b5R). X-ray diffraction data sets of HiPIP and b5R were also collected using the same crystals used for the neutron diffraction experiment. We have performed X-ray and neutron joint refinement, and we determined protonation states of amino acid residues and orientation of water molecules on the surface of the proteins.
resolution (b5R). X-ray diffraction data sets of HiPIP and b5R were also collected using the same crystals used for the neutron diffraction experiment. We have performed X-ray and neutron joint refinement, and we determined protonation states of amino acid residues and orientation of water molecules on the surface of the proteins.
 reductase aimed to neutron crystallography
reductase aimed to neutron crystallographyShoyama, Yoshinari; Tamada, Taro; Kuroki, Ryota; Kimura, Shigenobu*; Takeda, Kazuki*; Hayashi, Takuro*; Miki, Kunio*
no journal, ,
NADH-cytochrome 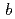
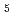 reductase (b5R) is a pyridine nucleotide-dependent flavin reductase which contains a FAD in a molecule. The b5R catalyzes the electron transfer from NADH to cytochrome b5. The b5R is related to fatty acid metabolism and reduction of cytochrome P450. In order to clarify detailed structure of b5R, we carried out purification of b5R from E coli cell according to the method previously reported. Since a large-size crystal is necessary for the neutron structure analysis. We addressed enlargement of crystal by macroseeding method and periodical addition of protein sample to the crystallization solution. We succeeded in a preparation of large-scale crystal with volume of 4 mm
reductase (b5R) is a pyridine nucleotide-dependent flavin reductase which contains a FAD in a molecule. The b5R catalyzes the electron transfer from NADH to cytochrome b5. The b5R is related to fatty acid metabolism and reduction of cytochrome P450. In order to clarify detailed structure of b5R, we carried out purification of b5R from E coli cell according to the method previously reported. Since a large-size crystal is necessary for the neutron structure analysis. We addressed enlargement of crystal by macroseeding method and periodical addition of protein sample to the crystallization solution. We succeeded in a preparation of large-scale crystal with volume of 4 mm , and we did preliminary neutron diffraction study using this crystal. Then we got 2
, and we did preliminary neutron diffraction study using this crystal. Then we got 2 resolution neutron diffraction data at BIX4.
resolution neutron diffraction data at BIX4.
Hirano, Yu; Tamada, Taro; Kurihara, Kazuo; Kusaka, Katsuhiro*; Miki, Kunio*
no journal, ,
Information about hydrogen atoms are important for understanding both structure and function of proteins. The structure of hydrogen atoms in proteins has been discussed based on the ideal values of bond lengths and angles obtained from structures of small molecules. A huge contribution to the protein science will be made by the structure information of hydrogen atoms without restraints of the ideal values. Hydrogen atoms are easily detected in neutron crystal structures because neutrons have a strong diffraction power compared to other atoms in proteins. In this study, we have determined high-resolution neutron structure of the high-potential iron-sulfur protein (HiPIP). The neutron diffraction experiment was performed at the BL03 (iBIX) beamline of J-PARC/MLF. We have obtained the diffraction data set at 1.1 -resolution that is the highest resolution data in protein neutron studies. The protonation states of amino acid residues at the surface of HiPIP have been observed after structure refinement. In addition, many deviations from the ideal values were observed in the distances and angles involved in hydrogen atoms.
-resolution that is the highest resolution data in protein neutron studies. The protonation states of amino acid residues at the surface of HiPIP have been observed after structure refinement. In addition, many deviations from the ideal values were observed in the distances and angles involved in hydrogen atoms.
Shoyama, Yoshinari; Tamada, Taro; Kuroki, Ryota; Kimura, Shigenobu*; Takeda, Kazuki*; Hayashi, Takuro*; Miki, Kunio*
no journal, ,
NADH-cytochrome b5 reductase (b5R) is a pyridine nucleotide-dependent flavin reductase which contains a FAD in a molecule. The b5R catalyzes the electron transfer from NADH to cytochrome b5. The b5R is related to fatty acid metabolism and reduction of cytochrome P450. In order to clarify detailed structure of b5R, we carried out purification of b5R from E coli cell according to the method previously reported. Since a large-size crystal is necessary for the neutron structure analysis, we addressed enlargement of crystal by macroseeding method and periodical addition of protein sample to the crystallization solution. We succeeded in a preparation of large-scale crystal with a size of 1.0 0.5
0.5 0.3 mm, which is suitable for preliminary neutron diffraction study.
0.3 mm, which is suitable for preliminary neutron diffraction study.
 reductase
reductaseYamada, Mitsugu; Tamada, Taro; Matsumoto, Fumiko; Takeda, Kazuki*; Kimura, Shigenobu*; Kuroki, Ryota; Miki, Kunio*
no journal, ,
no abstracts in English
Yamada, Mitsugu; Tamada, Taro; Matsumoto, Fumiko; Takeda, Kazuki*; Kimura, Shigenobu*; Kuroki, Ryota; Miki, Kunio*
no journal, ,
no abstracts in English
 reductase
reductaseYamada, Mitsugu; Tamada, Taro; Matsumoto, Fumiko; Takeda, Kazuki*; Kimura, Shigenobu*; Kuroki, Ryota; Miki, Kunio*
no journal, ,
NADH-cytochrome  reductase (b5R; EC 1.6.2.2) is a flavoprotein and catalyses two-electron transfer from NADH to cytochrome
reductase (b5R; EC 1.6.2.2) is a flavoprotein and catalyses two-electron transfer from NADH to cytochrome  through FAD. Here, we present crystal structures of fully reduced and two re-oxidized forms of b5Rs determined. The crystal structure of fully reduced b5R showed that the relative location of two domains slightly shifted in comparison with that of oxidized form. This shift allowed to create a new hydrogen bonding interaction between N5 atom of isoalloxazine ring and hydroxyl oxygen atom of Thr66. The isoalloxazine ring of FAD in fully reduced form is flat and stacked with the nicotinamide ring of bound NAD
through FAD. Here, we present crystal structures of fully reduced and two re-oxidized forms of b5Rs determined. The crystal structure of fully reduced b5R showed that the relative location of two domains slightly shifted in comparison with that of oxidized form. This shift allowed to create a new hydrogen bonding interaction between N5 atom of isoalloxazine ring and hydroxyl oxygen atom of Thr66. The isoalloxazine ring of FAD in fully reduced form is flat and stacked with the nicotinamide ring of bound NAD . Moreover, two re-oxidized forms (form-1 and 2) were prepared using fully reduced crystals by exposure to the air. The electron densities for the nicotinamide ring of NAD
. Moreover, two re-oxidized forms (form-1 and 2) were prepared using fully reduced crystals by exposure to the air. The electron densities for the nicotinamide ring of NAD was ambiguous in form-1 and was completely disappeared in form-2. These results suggested that re-oxidization follows a two-step mechanism in which nicotinamide moiety is released firstly and then whole NAD
was ambiguous in form-1 and was completely disappeared in form-2. These results suggested that re-oxidization follows a two-step mechanism in which nicotinamide moiety is released firstly and then whole NAD is released.
is released.
Hirano, Yu; Takeda, Kazuki*; Kurihara, Kazuo; Tamada, Taro; Miki, Kunio*
no journal, ,
High-potential iron-sulfur protein (HiPIP) is a soluble electron carrier protein that transfers electrons from cytochrome 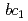 complex to the reaction center complex in photosynthetic purple bacteria. We prepared large crystals and examined conditions for diffraction data collection in order to obtain high-resolution neutron structure. We successfully obtained crystals with the volume over 2 mm
complex to the reaction center complex in photosynthetic purple bacteria. We prepared large crystals and examined conditions for diffraction data collection in order to obtain high-resolution neutron structure. We successfully obtained crystals with the volume over 2 mm by macro-seeding method. Preliminary diffraction analysis was performed at J-PARC, and the HiPIP crystal diffracted to 1.1
by macro-seeding method. Preliminary diffraction analysis was performed at J-PARC, and the HiPIP crystal diffracted to 1.1 resolution that is the highest resolution in the protein neutron diffraction data.
resolution that is the highest resolution in the protein neutron diffraction data.
Hirano, Yu; Tamada, Taro; Kurihara, Kazuo; Kusaka, Katsuhiro*; Ono, Hiraku*; Takeda, Kazuki*; Miki, Kunio*
no journal, ,
Hydrogen atoms are involved in protein folding and enzymatic reaction. Structures of hydrogen atoms in proteins have been discussed based on the ideal bond distances and angles obtained by small molecular crystallography. However, structural information about hydrogen atoms without geometric restraints is important for understanding structures and functions of proteins. High-potential iron-sulfur protein (HiPIP) is an electron carrier protein which functions in photosynthetic electron transfer chain of purple bacteria. In this work, we determined high-resolution neutron structure of HiPIP. The neutron diffraction experiment was performed at the BL03 beamline (iBIX) of J-PARC/MLF. We have collected the highest resolution data at 1.1 angstrom in the protein neutron structures. After structure refinement, we have observed many deviations in positions and bond lengths of hydrogen atoms from the ideal geometries.
 reductase by X-ray crystallography
reductase by X-ray crystallographyTamada, Taro; Yamada, Mitsugu*; Takeda, Kazuki*; Matsumoto, Fumiko*; Kimura, Shigenobu*; Kuroki, Ryota; Miki, Kunio*
no journal, ,
NADH-cytochrome  reductase (b5R) is a flavoprotein consisting of NADH- and FAD- domains, and catalyzes the electron transfer from two electron carriers of NADH to one electron carrier of cytochrome
reductase (b5R) is a flavoprotein consisting of NADH- and FAD- domains, and catalyzes the electron transfer from two electron carriers of NADH to one electron carrier of cytochrome  (Cb5). The structure of the fully reduced form of porcine liver b5R was determined using a crystal grown under anaerobic condition. In the reduced b5R structure, which was determined at 1.68
(Cb5). The structure of the fully reduced form of porcine liver b5R was determined using a crystal grown under anaerobic condition. In the reduced b5R structure, which was determined at 1.68  resolution, the relative location of the two domains was slightly shifted in comparison with that of the oxidized form. This shift resulted in an increase in the solvent accessible surface area of FAD, and created a new hydrogen bonding interaction between the N5 atom of the isoalloxazine ring of FAD and the hydroxyl oxygen atom of Thr66, which is considered to be a key residue in the release of a proton from the N5 atom. The isoalloxazine ring of FAD in the reduced form is flat, which is similar to that in the oxidized form, and is stacked with the nicotinamide ring of NAD
resolution, the relative location of the two domains was slightly shifted in comparison with that of the oxidized form. This shift resulted in an increase in the solvent accessible surface area of FAD, and created a new hydrogen bonding interaction between the N5 atom of the isoalloxazine ring of FAD and the hydroxyl oxygen atom of Thr66, which is considered to be a key residue in the release of a proton from the N5 atom. The isoalloxazine ring of FAD in the reduced form is flat, which is similar to that in the oxidized form, and is stacked with the nicotinamide ring of NAD . Both of reduced and oxidized b5R structures could explain how prevents the backflow and accelerates the transfer of electrons to one-electron acceptors, such as Cb5, in the catalytic cycle. Furthermore, the crystallographic analysis by the cryo-trapping method, which controls the exposure time of the fully reduced crystals against the air, suggested that re-oxidation follows a two-step mechanism. These results provide structural insights into the catalytic cycle of b5R.
. Both of reduced and oxidized b5R structures could explain how prevents the backflow and accelerates the transfer of electrons to one-electron acceptors, such as Cb5, in the catalytic cycle. Furthermore, the crystallographic analysis by the cryo-trapping method, which controls the exposure time of the fully reduced crystals against the air, suggested that re-oxidation follows a two-step mechanism. These results provide structural insights into the catalytic cycle of b5R.
Hirano, Yu; Kurihara, Kazuo; Kusaka, Katsuhiro*; Miki, Kunio*; Tamada, Taro
no journal, ,
Most of electron transfer proteins possess prosthetic groups, such as heme, flavin and iron-sulfur cluster, and electron transfer reaction is mediated by hydrogen atoms or valence shell electrons of the prosthetic groups. Therefore, structural information including hydrogen atoms is required for understanding their electron transfer mechanisms. We have performed high-resolution neutron structure analyses with 2 electron transfer proteins of high-potential iron-sulfur protein (HiPIP) and NADH-cytochrome 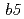 reductase (b5R). Diffraction experiments were performed at BL03 (iBIX) beamline of J-PARC using large crystals of HiPIP and b5R. Diffraction spots were observed up to 1.17
reductase (b5R). Diffraction experiments were performed at BL03 (iBIX) beamline of J-PARC using large crystals of HiPIP and b5R. Diffraction spots were observed up to 1.17  and 1.37
and 1.37  resolutions for HiPIP and b5R, respectively.
resolutions for HiPIP and b5R, respectively.
Hirano, Yu; Tamada, Taro; Kurihara, Kazuo; Kusaka, Katsuhiro*; Miki, Kunio*
no journal, ,
The redox states of high-potential iron-sulfur protein (HiPIP) have been reported to be stabilized by solvent molecules around the protein surface. Information about hydrogen atoms is required to determine the orientation of solvent molecules, especially that of water molecules. We have performed neutron diffraction experiment of HiPIP at iBIX beamline of J-PARC to obtain structural information about hydrogen atoms. The crystal of the oxidized form of HiPIP was obtained by macro-seeding method and the size of the crystal was reached to 1 mm . The crystal was soaked in deuterated solution for 3 months, and then the crystal was transferred to cryoprotectant solution for data collection at 100 K. The crystal was flash frozen in a nitrogen-gas stream at 100 K and it was cooled during the data collection. Diffraction spots were observed up to 1.1
. The crystal was soaked in deuterated solution for 3 months, and then the crystal was transferred to cryoprotectant solution for data collection at 100 K. The crystal was flash frozen in a nitrogen-gas stream at 100 K and it was cooled during the data collection. Diffraction spots were observed up to 1.1  resolution after exposure to neutrons for 9 hours.
resolution after exposure to neutrons for 9 hours.