Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Yamaguchi, Ko*; Kawaguchi, Daisuke*; Miyata, Noboru*; Miyazaki, Tsukasa*; Aoki, Hiroyuki; Yamamoto, Satoru*; Tanaka, Keiji*
Physical Chemistry Chemical Physics, 24(36), p.21578 - 21582, 2022/09
Times Cited Count:5 Percentile:70.33(Chemistry, Physical)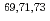 Co
CoLokotko, T.*; Leblond, S.*; Lee, J.*; Doornenbal, P.*; Obertelli, A.*; Poves, A.*; Nowacki, F.*; Ogata, Kazuyuki*; Yoshida, Kazuki; Authelet, G.*; et al.
Physical Review C, 101(3), p.034314_1 - 034314_7, 2020/03
Times Cited Count:10 Percentile:69.78(Physics, Nuclear)The structures of the neutron-rich 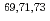 Co isotopes were investigated via (
Co isotopes were investigated via (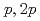 ) knockout reactions at the Radioactive Isotope Beam Factory, RIKEN. Level schemes were reconstructed using the
) knockout reactions at the Radioactive Isotope Beam Factory, RIKEN. Level schemes were reconstructed using the 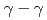 coincidence technique, with tentative spin-parity assignments based on the measured inclusive and exclusive cross sections. Comparison with shell-model calculations suggests coexistence of spherical and deformed shapes at low excitation energies in the
coincidence technique, with tentative spin-parity assignments based on the measured inclusive and exclusive cross sections. Comparison with shell-model calculations suggests coexistence of spherical and deformed shapes at low excitation energies in the 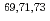 Co isotopes.
Co isotopes.
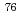 Ni from the (
Ni from the (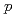 ,
,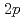 ) reaction
) reactionElekes, Z.*; Kripk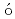 ,
, 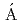 *; Sohler, D.*; Sieja, K.*; Ogata, Kazuyuki*; Yoshida, Kazuki; Doornenbal, P.*; Obertelli, A.*; Authelet, G.*; Baba, Hidetada*; et al.
*; Sohler, D.*; Sieja, K.*; Ogata, Kazuyuki*; Yoshida, Kazuki; Doornenbal, P.*; Obertelli, A.*; Authelet, G.*; Baba, Hidetada*; et al.
Physical Review C, 99(1), p.014312_1 - 014312_7, 2019/01
Times Cited Count:10 Percentile:64.15(Physics, Nuclear)The nuclear structure of the 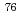 Ni nucleus was investigated by (
Ni nucleus was investigated by (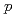 ,
,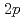 ) reaction using a NaI(Tl) array to detect the deexciting prompt
) reaction using a NaI(Tl) array to detect the deexciting prompt  rays. A new transition with an energy of 2227 keV was identified by
rays. A new transition with an energy of 2227 keV was identified by 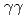 and
and 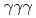 coincidences. Our shell-model calculations using the Lenzi, Nowacki, Poves, and Sieja interaction produced good candidates for the experimental proton hole states in the observed energy region, and the theoretical cross sections showed good agreement with the experimental values. Although we could not assign all the experimental states to the theoretical ones unambiguously, the results are consistent with a reasonably large Z = 28 shell gap for nickel isotopes in accordance with previous studies.
coincidences. Our shell-model calculations using the Lenzi, Nowacki, Poves, and Sieja interaction produced good candidates for the experimental proton hole states in the observed energy region, and the theoretical cross sections showed good agreement with the experimental values. Although we could not assign all the experimental states to the theoretical ones unambiguously, the results are consistent with a reasonably large Z = 28 shell gap for nickel isotopes in accordance with previous studies.
Toyomori, Yuka*; Tsuji, Satoru*; Mitsuda, Shinobu*; Okayama, Yoichi*; Ashida, Shiomi*; Mori, Atsunori*; Kobayashi, Toru; Miyazaki, Yuji; Yaita, Tsuyoshi; Arae, Sachie*; et al.
Bulletin of the Chemical Society of Japan, 89(12), p.1480 - 1486, 2016/09
Times Cited Count:8 Percentile:29.47(Chemistry, Multidisciplinary)Hiraishi, Masatoshi*; Iimura, Soshi*; Kojima, Kenji*; Yamaura, Junichi*; Hiraka, Haruhiro*; Ikeda, Kazutaka*; Miao, P.*; Ishikawa, Yoshihisa*; Torii, Shuki*; Miyazaki, Masanori*; et al.
Nature Physics, 10(4), p.300 - 303, 2014/04
Times Cited Count:103 Percentile:95.46(Physics, Multidisciplinary)Yamasaki, Chisato*; Murakami, Katsuhiko*; Fujii, Yasuyuki*; Sato, Yoshiharu*; Harada, Erimi*; Takeda, Junichi*; Taniya, Takayuki*; Sakate, Ryuichi*; Kikugawa, Shingo*; Shimada, Makoto*; et al.
Nucleic Acids Research, 36(Database), p.D793 - D799, 2008/01
Times Cited Count:51 Percentile:71.25(Biochemistry & Molecular Biology)Here we report the new features and improvements in our latest release of the H-Invitational Database, a comprehensive annotation resource for human genes and transcripts. H-InvDB, originally developed as an integrated database of the human transcriptome based on extensive annotation of large sets of fulllength cDNA (FLcDNA) clones, now provides annotation for 120 558 human mRNAs extracted from the International Nucleotide Sequence Databases (INSD), in addition to 54 978 human FLcDNAs, in the latest release H-InvDB. We mapped those human transcripts onto the human genome sequences (NCBI build 36.1) and determined 34 699 human gene clusters, which could define 34 057 protein-coding and 642 non-protein-coding loci; 858 transcribed loci overlapped with predicted pseudogenes.