Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Tominaga, Taiki*; Nakagawa, Hiroshi; Sahara, Masae*; Oda, Takashi*; Inoue, Rintaro*; Sugiyama, Masaaki*
Life (Internet), 12(5), p.675_1 - 675_9, 2022/05
Times Cited Count:0 Percentile:0.01(Biology)The background scattering of sample cells suitable for aqueous protein solution samples, conducted with a neutron backscattering spectrometer, was evaluated. It was found that the scattering intensity of an aluminum sample cell coated with boehmite using D O was lower than that of a sample cell coated with regular water (H
O was lower than that of a sample cell coated with regular water (H O). In addition, meticulous attention to cells with small individual weight differences and the positional reproducibility of the sample cell relative to the spectrometer neutron beam position enabled the accurate subtraction of the scattering profiles of the D
O). In addition, meticulous attention to cells with small individual weight differences and the positional reproducibility of the sample cell relative to the spectrometer neutron beam position enabled the accurate subtraction of the scattering profiles of the D O buffer and the sample container. Consequently, high quality information on protein dynamics could be extracted from dilute protein solutions.
O buffer and the sample container. Consequently, high quality information on protein dynamics could be extracted from dilute protein solutions.
Nakagawa, Hiroshi; Saio, Tomohide*; Nagao, Michihiro*; Inoue, Rintaro*; Sugiyama, Masaaki*; Ajito, Satoshi; Tominaga, Taiki*; Kawakita, Yukinobu
Biophysical Journal, 120(16), p.3341 - 3354, 2021/08
Times Cited Count:4 Percentile:38.66(Biophysics)A multi-domain protein can have various conformations in solution. Interactions with other molecules result in the stabilization of one of the conformations and change in the domain dynamics. SAXS, a well-established experimental technique, can be employed to elucidate the conformation of a multi-domain protein in solution. NSE spectroscopy is a promising technique for recording the domain dynamics in nanosecond and nanometer scale. Despite the great efforts, there are still under development. Thus, we quantitatively removed the contribution of diffusion dynamics and hydrodynamic interactions from the NSE data via incoherent scattering, revealing the differences in the domain dynamics of the three functional states of a multi-domain protein, MurD. The differences among the three states can be explained by two domain modes.
Inoue, Rintaro*; Oda, Takashi*; Nakagawa, Hiroshi; Tominaga, Taiki*; Saio, Tomohide*; Kawakita, Yukinobu; Shimizu, Masahiro*; Okuda, Aya*; Morishima, Ken*; Sato, Nobuhiro*; et al.
Scientific Reports (Internet), 10, p.21678_1 - 21678_10, 2020/12
Times Cited Count:3 Percentile:13.48(Multidisciplinary Sciences)Incoherent quasielastic neutron scattering (iQENS) is a fascinating technique for investigating the internal dynamics of protein. However, both low flux of neutron beam and absence of analytical procedure for extracting the internal dynamics from iQENS profile have been obstacles for studying it under physiological condition (in solution). Thanks to the recent development of neutron source, spectrometer and computational technique, they enable us to decouple internal dynamics, translational and rotational diffusions from the iQENS profile. The internal dynamics of two proteins: globular domain protein (GDP) and intrinsically disordered protein (IDP) in solution were studied. It was found that the average relaxation rate of IDP was larger than that of GDP. Through the detailed analyses on their internal dynamics, it was revealed that the fraction of mobile H atoms in IDP was much higher than that in GDP. Interestingly, the fraction of mobile H atoms was closely related to the fraction of H atoms on highly solvent exposed surfaces. The iQENS study presented that the internal dynamics were governed by the highly solvent exposed amino acid residues depending upon protein molecular architectures.
Numata, Masaaki; Ogura, Yuichi*; Nakagawa, Yosuke; Inoue, Naoko
Dai-41-Kai Nihon Kaku Busshitsu Kanri Gakkai Nenji Taikai Kaigi Rombunshu (Internet), 3 Pages, 2020/11
JAEA/ISCN is actively using a virtual reality (VR) system for training in nuclear non-proliferation safeguards and nuclear security. The VR system allows participants to experience the environment of a nuclear power plant and a research reactor in a cyber space. It is beneficial for the participants who have never been in a nuclear power plant to see how they are even virtually, in order to learn their physical protection system. Workstations of OS for VR use Windows7 and the warrantee protection period expired in March 2020. For that reason, it was decided to renew those workstations over 2019-2020. This paper will describe its planning called "Refresh Project", which aims to minimize impacts on training and visitors, as well as first year achievement.
Sugiyama, Masaaki*; Inoue, Rintaro*; Nakagawa, Hiroshi; Saio, Tomohide*
Hamon, 30(1), p.16 - 25, 2020/02
Neutron has distinct features as a scattering probe to analyze structure and dynamics of biological macromolecules. The theme of this review is to try to describe how we did/do utilize them. And "How we should utilize them more effectively in the trend of integrative structural biology?" with solution scattering.
Sugiyama, Masaaki*; Nakagawa, Hiroshi; Inoue, Rintaro*; Kawakita, Yukinobu
JAEA-Review 2017-024, 40 Pages, 2017/12
Now-a-days, promotion of life science by utilizing neutron (neutrons biology) is highly demanded in our country, following installation and improvement of high quality and intensity neutron sources at J-PARC and JRR-3. Aiming at accelerating development of neutrons biology in our country, an international workshop "Neutron biology for next generation" was held as a J-PARC Workshop at Ibaraki Quantum Beam Research Center from 22 March to 23 March in 2017. In the workshop, latest instruments, new-fashioned methodologies, recent scientific results and future perspectives were extensively discussed by domestic neutron instrumental scientists and domestic/foreign neutron biologists. This is a report of the workshop summarized by organizers.
Kataoka, Takahiro*; Sakoda, Akihiro*; Yoshimoto, Masaaki*; Nakagawa, Shinya*; Toyota, Teruaki*; Nishiyama, Yuichi*; Yamato, Keiko*; Ishimori, Yuu; Kawabe, Atsushi*; Hanamoto, Katsumi*; et al.
Radiation Protection Dosimetry, 146(1-3), p.360 - 363, 2011/07
Times Cited Count:6 Percentile:44.28(Environmental Sciences)Our previous studies showed the possibility that activation of the antioxidative function alleviates various oxidative damages, which are related to lifestyle diseases. Results showed that, low-dose X-ray irradiation activated superoxide dismutase and inhibits oedema following ischaemia-reperfusion. To alleviate ischaemia-reperfusion injury with transplantation, the changes of the antioxidative function in liver graft using low-dose X-ray irradiation immediately after exenteration were examined. Results showed that liver grafts activate the antioxidative function as a result of irradiation. In addition, radon inhalation enhances the antioxidative function in some organs, and alleviates alcohol-induced oxidative damage of mouse liver. Moreover, in order to determine the most effective condition of radon inhalation, mice inhaled radon before or after carbon tetrachloride (CCl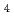 ) administration. Results showed that radon inhalation alleviates CCl
) administration. Results showed that radon inhalation alleviates CCl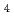 -induced hepatopathy, especially prior inhalation. It is highly possible that adequate activation of antioxidative functions induced by low-dose irradiation can contribute to preventing or reducing oxidative damages, which are related to lifestyle diseases.
-induced hepatopathy, especially prior inhalation. It is highly possible that adequate activation of antioxidative functions induced by low-dose irradiation can contribute to preventing or reducing oxidative damages, which are related to lifestyle diseases.
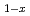 LaCe
LaCe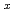 CuO
CuO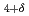 cuprate superconductors
cuprate superconductorsFujita, Masaki*; Matsuda, Masaaki; Lee, S.-H.*; Nakagawa, Masaaki*; Yamada, Kazuyoshi*
Physical Review Letters, 101(10), p.107003_1 - 107003_4, 2008/09
Times Cited Count:37 Percentile:80.76(Physics, Multidisciplinary)Low-energy spin fluctuations have been investigated in the electron-doped Pr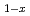 LaCe
LaCe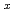 CuO
CuO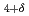 over a wide concentration range of 0.07
over a wide concentration range of 0.07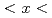 0.18, spanning from the antiferromagnetic phase to the heavily overdoped superconducting (SC) phase. The low-energy excitations exhibit commensurate peaks centered at the (
0.18, spanning from the antiferromagnetic phase to the heavily overdoped superconducting (SC) phase. The low-energy excitations exhibit commensurate peaks centered at the (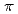 ,
, 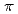 ) position for all
) position for all 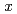 . Our data show that the characteristics of the excitations, such as the relaxation rate and the spin stiffness, decrease with increasing
. Our data show that the characteristics of the excitations, such as the relaxation rate and the spin stiffness, decrease with increasing 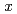 in the SC phase and disappear with the disappearance of superconductivity.
in the SC phase and disappear with the disappearance of superconductivity.
Nagashima, Keisuke; Kikuchi, Mitsuru; Kurita, Genichi; Ozeki, Takahisa; Aoyagi, Tetsuo; Ushigusa, Kenkichi; Neyatani, Yuzuru; Kubo, Hirotaka; Mori, Kensuke*; Nakagawa, Shoji*; et al.
Fusion Engineering and Design, 36(2-3), p.325 - 342, 1997/05
Times Cited Count:1 Percentile:14.48(Nuclear Science & Technology)no abstracts in English
Sato, Junya; Suzuki, Shinji; Nakagawa, Akinori; Kato, Jun; Sakakibara, Tetsuro; Nakazawa, Osamu; Yamashita, Masaaki; Sato, Fuminori; Sukegawa, Hirobumi; Meguro, Yoshihiro
no journal, ,
no abstracts in English
Nakagawa, Hiroshi; Saio, Tomohide*; Sugiyama, Masaaki*; Inoue, Rintaro*; Nagao, Michihiro*
no journal, ,
It is necessary for the understanding of structure polymorphism of various molecules and interacting protein and the plastic molecules base to clarify the fluctuation of the domain as the mer. In this study, I targeted protein MurD consisting of three domains and a ligand was in a free state and analyzed domain exercise by the quantum beam dispersion method using X-rays and the neutron and the correlation structure analytical method that fused of the molecules simulation about an ATP-binding state, three states of the Compound1-binding state. By the solution small angle scattering experiment, I showed that the solution structure of the 3 state was different. In addition, I showed good agreement with the dispersion profile obtained from the molecular simulation and confirmed that I could discuss solution structure in atom resolving power from experimental data of the low resolving power. I extracted a domain campaign in conjunction with the function from the chief ingredient analysis of the molecular simulation result. Furthermore, a result to suggest what a fluctuation and a couple of the amino acid residue that this domain campaign participated in the combination with interaction molecules of MurD did was provided. By the method that fused by an experiment method of dynamic solution structure analysis including a neutron spin echo and technique of the calculation science in addition to small angle scattering, I visualize domain exercise of protein in atom resolving power and, in the announcement, argue by a function of the protein from the dynamic structure that a couple did between the hierarchies of different space scale.
Nakagawa, Hiroshi; Saio, Tomohide*; Sugiyama, Masaaki*; Inoue, Rintaro*
no journal, ,
It is necessary to check a structural change of the protein on the domain scale to predict the interaction with the target molecule and interaction between the protein based on tertiary structure information of the protein at the atom level. In addition, it is necessary for the understanding of structure polymorphism of various molecules and interacting protein and the plastic molecules base to clarify the fluctuation of the domain as the mer. It becomes the important problem how you elucidate flexibility of such a protein structure in the next-generation structural biology. In this study, I analyze a change of the multi-domain protein structure by the quantum beam dispersion method using X-rays and the neutron and the correlation structure analytical method that fused of the molecular simulation. In addition, I analyze the local structure of the active site of the protein in conjunction with a domain structure by quoting molecular simulation. I mediate between a solution dispersion experiment of the low resolving power and two experiment information of the crystal structure of the atom resolving power that has been already untied by a computer technology and elucidate the protein interaction that plural domains weave in wide space resolving power to be able to foresee the whole complex from an atom level.
Nakagawa, Hiroshi; Saio, Tomohide*; Sugiyama, Masaaki*; Inoue, Rintaro*; Tominaga, Taiki*
no journal, ,
The neutron associate elastic scattering device installed in large strength pulse neutron J-PARC is effective for the analysis of the protein dynamics of the pico second - nanosecond. It will show the importance of the QENS experiment of the protein using J-PARC to clarify how the dynamics of the space scale are related with cooperative dynamics of the whole protein affecting creature function expression then. On the other hand, for the structural biology to discuss a function based on a tertiary structure, it is difficult to relate a function to structure dynamics only from QENS spectrum. It is effective to quote molecular simulation so that structure discusses the information of dynamics to relate a function to structure scientifically. I suggest that I visualize the hierarchical structure of protein dynamics in atom resolving power by the MD-Neutron method which fused by a neutron dispersion experiment and molecules simulation while utilizing various technique of the structural biology in this announcement from different angles. In addition, about MurD which is multi-domain protein, I discuss a function of the protein from the dynamic structure that a couple did between the hierarchies of the scale between the different space-time.
Nakagawa, Hiroshi; Saio, Tomohide*; Oda, Takashi*; Sato, Mamoru*; Inoue, Rintaro*; Sugiyama, Masaaki*; Tominaga, Taiki*; Kawakita, Yukinobu
no journal, ,
Protein is thermally fluctuating in solution, and the dynamics is essential for its biological functions. A protein has hierarchal structure and dynamics in temporal and spatial scale. In this work, QENS of a multi-domain protein, MurD, were measured by the TOF near backscattering spectrometer DNA in order to observe the internal motions. Hef is classified as an intrinsically disordered protein, which lost the rigid folded domain structure, and then have a more flexible structure than a folded protein. We will discuss the data treatment and analytical method of QENS of protein solutions, and characteristic dynamical features of folded rigid and disordered flexible proteins in the presentation.
Nakagawa, Hiroshi; Inoue, Rintaro*; Oda, Takashi*; Yagi, Maho*; Saio, Tomohide*; Oroguchi, Tomotaka*; Nagata, Yuya*; Sugiyama, Masaaki*; Sato, Mamoru*; Kawakita, Yukinobu; et al.
no journal, ,
Proteins have hierarchical structures and hierarchical dynamics. The importance of analyzing the structural dynamics of proteins in solution has been increasing in recent years, and neutron scattering is expected to be an experimental method to study the hierarchical structures on the nanometer order and the dynamics on the pico- to nanosecond time scales, which are relevant to biological functions. In this study, we used the quasi-elastic neutron scattering and small-angle neutron scattering instruments installed at MLF to elucidate the dynamics of these structures in space and time. We aim to establish a new generation of neutron structural biology that will lead protein science in Japan.
Nakagawa, Hiroshi; Saio, Tomohide*; Nagao, Michihiro*; Inoue, Rintaro*; Sugiyama, Masaaki*; Tominaga, Taiki*; Kawakita, Yukinobu
no journal, ,
The flexible conformation of a multidomain protein is responsible for its biological function. 3-domain protein: MurD (47kDa) changes its domain conformation sequentially from open to semi-closed to closed conformation during enzymatic reactions. However, the dynamics of the domains in each conformation is unknown. In this study, we combined small-angle X-ray and neutron scattering (SAXS and SANS), dynamic light scattering (DLS), neutron back scattering (NBS), neutron spin echo (NSE), and molecular dynamics (MD) simulations to investigate the conformational dynamics of MurD in the three corresponding states (apo and ATP, inhibitor-bound state). The analysis showed that the conformational dynamics of MurD during the enzymatic reaction was not affected. The analysis suggests that the changes in domain dynamics during enzymatic reactions are related to the affinity and reaction efficiency with ligands that bind specifically to each reaction state.
Nakagawa, Hiroshi; Saio, Tomohide*; Nagao, Michihiro*; Inoue, Rintaro*; Sugiyama, Masaaki*; Tominaga, Taiki*; Kawakita, Yukinobu
no journal, ,
The flexible conformation of a multidomain protein is responsible for its biological function. 3-domain protein: MurD (47kDa) changes its domain conformation sequentially from open to semi-closed to closed conformation during enzymatic reactions. However, the dynamics of the domains in each conformation is unknown. In this study, we combined small-angle X-ray and neutron scattering (SAXS and SANS), dynamic light scattering (DLS), neutron back scattering (NBS), neutron spin echo (NSE), and molecular dynamics (MD) simulations to investigate the conformational dynamics of MurD in the three corresponding states (apo and ATP, inhibitor-bound state). The analysis showed that the conformational dynamics of MurD during the enzymatic reaction was not affected. The analysis suggests that the changes in domain dynamics during enzymatic reactions are related to the affinity and reaction efficiency with ligands that bind specifically to each reaction state.
Nakagawa, Hiroshi; Inoue, Rintaro*; Oda, Takashi*; Yagi-Utsumi, Maho*; Saio, Tomohide*; Oroguchi, Tomotaka*; Nagata, Yuya*; Sugiyama, Masaaki*; Sato, Mamoru*; Kawakita, Yukinobu; et al.
no journal, ,
Proteins have hierarchical structures and hierarchical dynamics. Neutron scattering is expected to be a promising experimental technique to study the hierarchical structure on the nanometer order related to biological functions and the structure and dynamics on the pico- to nanosecond time scale. The research was initiated with the aim of elucidating the structure and dynamics of these spatiotemporal scales (MLF Long-term Proposal 2019L0300). In this proposal, we used samples of characterized functional multidomain proteins, including folded proteins and naturally denatured proteins, to promote our research. Researchers in a wide range of specialized fields collaborated to develop "a protein partial deuteration method utilizing the features of neutron scattering", "a wide spatio-temporal seamless analysis method by linking various neutron spectrometers", "a method for visualizing protein dynamics by linking neutron and computational science," and "a neutron quasi-elastic scattering measurement method for dilute protein solutions" and to We have pioneered a new generation of neutron structural biology that will lead protein science in Japan.