Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
 -glucanase from
-glucanase from 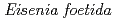
Arimori, Takao*; Ito, Akihiro*; Nakazawa, Masami*; Ueda, Mitsuhiro*; Tamada, Taro
Journal of Synchrotron Radiation, 20(6), p.884 - 889, 2013/11
Times Cited Count:17 Percentile:65.54(Instruments & Instrumentation)The saccharification process is essential for bioethanol production from woody biomass including celluloses. Cold-adapted cellulase, which has sufficient activity at low temperature (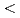 293 K), is capable of reducing heating costs during the saccharification process and is suitable for simultaneous saccharification and fermentation. Endo-1,4-
293 K), is capable of reducing heating costs during the saccharification process and is suitable for simultaneous saccharification and fermentation. Endo-1,4- -glucanase from the earthworm Eisenia fetida (EF-EG2) belonging to glycoside hydrolase family 9 has been shown to have the highest activity at 313 K, and also retained a comparatively high activity at 283 K. The recombinant EF-EG2 was purified expressed in Pichia pastoris, and then grew needle-shaped crystals with dimensions of 0.02
-glucanase from the earthworm Eisenia fetida (EF-EG2) belonging to glycoside hydrolase family 9 has been shown to have the highest activity at 313 K, and also retained a comparatively high activity at 283 K. The recombinant EF-EG2 was purified expressed in Pichia pastoris, and then grew needle-shaped crystals with dimensions of 0.02  0.02
0.02  1 mm. The crystals belonged to the space group P3221 with unit-cell parameters of
1 mm. The crystals belonged to the space group P3221 with unit-cell parameters of 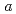 =
= 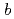 =136
=136  ,
, 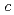 = 55.0
= 55.0  ,. The final model of EF-EG2, including 435 residues, two ions, seven crystallization reagents and 696 waters, was refined to a crystallographic
,. The final model of EF-EG2, including 435 residues, two ions, seven crystallization reagents and 696 waters, was refined to a crystallographic 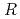 -factor of 14.7% (free
-factor of 14.7% (free 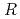 -factor of 16.8%) to 1.5
-factor of 16.8%) to 1.5  , resolution. The overall structure of EF-EG2 has an (
, resolution. The overall structure of EF-EG2 has an ( /
/ )
)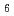 barrel fold which contains a putative active-site cleft and a negatively charged surface. This structural information helps us understand the catalytic and cold adaptation mechanisms of EF-EG2.
barrel fold which contains a putative active-site cleft and a negatively charged surface. This structural information helps us understand the catalytic and cold adaptation mechanisms of EF-EG2.
 sp. A-471 reveals a unique arrangement of the catalytic residues for inverting chitin hydrolysis
sp. A-471 reveals a unique arrangement of the catalytic residues for inverting chitin hydrolysisArimori, Takao*; Kawamoto, Noriko*; Shinya, Shoko*; Okazaki, Nobuo*; Nakazawa, Masami*; Miyatake, Kazutaka*; Fukamizo, Tamo*; Ueda, Mitsuhiro*; Tamada, Taro
Journal of Biological Chemistry, 288(26), p.18696 - 18706, 2013/07
Times Cited Count:30 Percentile:64.37(Biochemistry & Molecular Biology)Chitinase C from  sp. A-471 (Ra-ChiC) has a catalytic domain sequence similar to goose type (G-type) lysozymes and, unlike other chitinases, belongs to glycohydrolase (GH) family 23. Using NMR spectroscopy, however, Ra-ChiC was found to interact only with the chitin dimer but not with the peptideglycan fragment. Here we report the crystal structures of wild-type, E141Q, and E162Q of the catalytic domain of Ra-ChiC with or without chitin oligosaccharides. Ra-ChiC has a substrate-binding site including a tunnel-shaped cavity, which determines the substrate specificity. Mutation analyses based on this structural information indicated that a highly conserved Glu141 acts as a catalytic acid, and that Asp226 located at the roof of the tunnel activates a water molecule as a catalytic base. The unique arrangement of the catalytic residues makes a clear contrast to the other GH23 members and also to inverting GH19 chitinases.
sp. A-471 (Ra-ChiC) has a catalytic domain sequence similar to goose type (G-type) lysozymes and, unlike other chitinases, belongs to glycohydrolase (GH) family 23. Using NMR spectroscopy, however, Ra-ChiC was found to interact only with the chitin dimer but not with the peptideglycan fragment. Here we report the crystal structures of wild-type, E141Q, and E162Q of the catalytic domain of Ra-ChiC with or without chitin oligosaccharides. Ra-ChiC has a substrate-binding site including a tunnel-shaped cavity, which determines the substrate specificity. Mutation analyses based on this structural information indicated that a highly conserved Glu141 acts as a catalytic acid, and that Asp226 located at the roof of the tunnel activates a water molecule as a catalytic base. The unique arrangement of the catalytic residues makes a clear contrast to the other GH23 members and also to inverting GH19 chitinases.
 sp. A-471
sp. A-471Okazaki, Nobuo; Arimori, Takao; Nakazawa, Masami*; Miyatake, Kazutaka*; Ueda, Mitsuhiro*; Tamada, Taro
Acta Crystallographica Section F, 67(4), p.494 - 497, 2011/04
Times Cited Count:3 Percentile:41.34(Biochemical Research Methods) sp. A-471
sp. A-471Tamada, Taro; Okazaki, Nobuo; Ueda, Mitsuhiro*; Nakazawa, Masami*; Miyatake, Kazutaka*; Kuroki, Ryota
no journal, ,
Crystal structure of a novel chitinase (Ra-ChiC), a member of GH family 23, from the moderately thermophilic bacterium 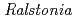 sp. A-471 has been solved in the active site to 1.9
sp. A-471 has been solved in the active site to 1.9  resolution. Crystal structure of Ra-ChiC was resemble to that of g-type lysozyme. It is well known that the residues involved in catalysis of the g-type lysozymes are Glu73, Asp90, and Asp101. The Glu73 (proton donor) in g-type lysozymes was conserved as Glu141 in the catalytic domain of Ra-ChiC.
resolution. Crystal structure of Ra-ChiC was resemble to that of g-type lysozyme. It is well known that the residues involved in catalysis of the g-type lysozymes are Glu73, Asp90, and Asp101. The Glu73 (proton donor) in g-type lysozymes was conserved as Glu141 in the catalytic domain of Ra-ChiC.
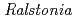 sp. A-471
sp. A-471Ueda, Mitsuhiro*; Nakazawa, Masami*; Miyatake, Kazutaka*; Okazaki, Nobuo; Kuroki, Ryota; Tamada, Taro
no journal, ,
Crystal structure of a novel chitinase (Ra-ChiC), a member of GH family 23, from the moderately thermophilic bacterium 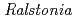 sp. A-471 has been solved in the active site to 1.9
sp. A-471 has been solved in the active site to 1.9  resolution. Crystal structure of Ra-ChiC was resemble to that of g-type lysozyme. It is well known that the residues involved in catalysis of the g-type lysozymes are Glu73, Asp90, and Asp101. The Glu73 (proton donor) in g-type lysozymes was conserved as Glu141 in the catalytic domain of Ra-ChiC.
resolution. Crystal structure of Ra-ChiC was resemble to that of g-type lysozyme. It is well known that the residues involved in catalysis of the g-type lysozymes are Glu73, Asp90, and Asp101. The Glu73 (proton donor) in g-type lysozymes was conserved as Glu141 in the catalytic domain of Ra-ChiC.
Arimori, Takao; Okazaki, Nobuo; Nakazawa, Masami*; Miyatake, Kazutaka*; Ueda, Mitsuhiro*; Tamada, Taro
no journal, ,
no abstracts in English
Arimori, Takao; Kawamoto, Noriko*; Okazaki, Nobuo; Nakazawa, Masami*; Miyatake, Kazutaka*; Ueda, Mitsuhiro*; Tamada, Taro
no journal, ,
no abstracts in English
 -glucanase from Eisenia foetida
-glucanase from Eisenia foetidaArimori, Takao*; Ito, Akihiro*; Nakazawa, Masami*; Ueda, Mitsuhiro*; Tamada, Taro
no journal, ,
 -endoglucanase from
-endoglucanase from 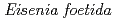
Arimori, Takao*; Ito, Akihiro*; Nakazawa, Masami*; Ueda, Mitsuhiro*; Tamada, Taro
no journal, ,
no abstracts in English
Arimori, Takao*; Kawamoto, Noriko*; Okazaki, Nobuo*; Nakazawa, Masami*; Miyatake, Kazutaka*; Fukamizo, Tamo*; Ueda, Mitsuhiro*; Tamada, Taro
no journal, ,
Chitin, linear  -1,4-linked polymer of
-1,4-linked polymer of 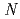 -acetyl-D-glucosamine (NAG), is the second abundant biopolymer in nature next to cellulose. Hydrolysis of chitin provides useful products,
-acetyl-D-glucosamine (NAG), is the second abundant biopolymer in nature next to cellulose. Hydrolysis of chitin provides useful products, 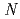 -acetyl-chitooligosaccharides [(NAG)
-acetyl-chitooligosaccharides [(NAG)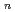 ] and chitooligosaccharides, which have a variety of biological functions including antibacterial activity and antitumor activity. We have previously cloned a novel chitinase gene from a moderate thermophilic strain
] and chitooligosaccharides, which have a variety of biological functions including antibacterial activity and antitumor activity. We have previously cloned a novel chitinase gene from a moderate thermophilic strain 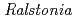 sp. A-471 (Ra-ChiC). Ra-ChiC comprises a signal peptide, a chitin-binding domain, an interdomain linker, and a catalytic domain. The catalytic domain shares amino acid sequence homology with goose type (G-type) lysozymes and, unlike other chitinases, Ra-ChiC belongs to glycohydrolase (GH) family 23. However, Ra-ChiC does not exhibit lysozyme activity, but only chitinase activity. In this study, we aim to reveal how Ra-ChiC catalyzes the hydrolysis of chitin and why Ra-ChiC exhibits chitinase activity instead of lysozyme activity. We determined the crystal structures of the catalytic domain of Ra-ChiC (Ra-ChiC
sp. A-471 (Ra-ChiC). Ra-ChiC comprises a signal peptide, a chitin-binding domain, an interdomain linker, and a catalytic domain. The catalytic domain shares amino acid sequence homology with goose type (G-type) lysozymes and, unlike other chitinases, Ra-ChiC belongs to glycohydrolase (GH) family 23. However, Ra-ChiC does not exhibit lysozyme activity, but only chitinase activity. In this study, we aim to reveal how Ra-ChiC catalyzes the hydrolysis of chitin and why Ra-ChiC exhibits chitinase activity instead of lysozyme activity. We determined the crystal structures of the catalytic domain of Ra-ChiC (Ra-ChiC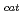 ), Ra-ChiC
), Ra-ChiC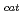 complexed with (NAG)
complexed with (NAG) , E141Q mutant of Ra-ChiC
, E141Q mutant of Ra-ChiC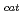 complexed with (NAG)
complexed with (NAG)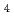 , E162Q mutant of Ra-ChiC
, E162Q mutant of Ra-ChiC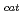 , and E162Q mutant of Ra-ChiC
, and E162Q mutant of Ra-ChiC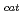 complexed with (NAG)
complexed with (NAG) . These structures provided us structural basis of substrate recognition mechanism and revealed that Ra-ChiC has a unique substrate-binding site including a tunnel-shaped cavity, which determines the substrate specificity. In addition, we also carried out a mutation analysis of acidic amino acid residues located at the active site. As a result, we found that not only a highly conserved Glu141 but also Asp226 located at the roof of the tunnel have quite important roles in catalysis.
. These structures provided us structural basis of substrate recognition mechanism and revealed that Ra-ChiC has a unique substrate-binding site including a tunnel-shaped cavity, which determines the substrate specificity. In addition, we also carried out a mutation analysis of acidic amino acid residues located at the active site. As a result, we found that not only a highly conserved Glu141 but also Asp226 located at the roof of the tunnel have quite important roles in catalysis.