Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
 is characterized as Type II bacterial topoisomerase and its activity is differentially regulated by PprA in vitro
is characterized as Type II bacterial topoisomerase and its activity is differentially regulated by PprA in vitroKota, S.*; Rajpurohit, Y. S.*; Charaka, V. K.*; Sato, Katsuya; Narumi, Issey*; Misra, H, S.*
Extremophiles, 20(2), p.195 - 205, 2016/03
Times Cited Count:12 Percentile:40.92(Biochemistry & Molecular Biology) by ion beam breeding technology
by ion beam breeding technologySato, Katsuya; Ueda, Ryoshiro*; Hase, Yoshihiro; Narumi, Issey*; Ono, Yutaka
JAEA-Review 2015-022, JAEA Takasaki Annual Report 2014, P. 100, 2016/02

Ueda, Ryoshiro*; Sato, Katsuya; Hayashi, Hidenori*; Narumi, Issey*; Ono, Yutaka
JAEA-Review 2015-022, JAEA Takasaki Annual Report 2014, P. 101, 2016/02
 mutant
mutantKitamura, Satoshi; Ono, Yutaka; Narumi, Issey*
JAEA-Review 2015-022, JAEA Takasaki Annual Report 2014, P. 64, 2016/02
Forward genetics approaches have helped elucidate the anthocyanin biosynthetic pathway in plants. We used the 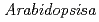 banyuls (ban) mutant, which accumulates anthocyanins, instead of colorless proanthocyanidin precursors, in immature seeds. In contrast to standard screens for mutants lacking anthocyanins in leaves/stems, we mutagenized ban plants and screened for mutants showing differences in pigmentation of immature seeds. The pale banyuls1 (pab1) mutation was found to show reduced levels of anthocyanins in immature seed coat. Here we characterize this mutant at the molecular level.
banyuls (ban) mutant, which accumulates anthocyanins, instead of colorless proanthocyanidin precursors, in immature seeds. In contrast to standard screens for mutants lacking anthocyanins in leaves/stems, we mutagenized ban plants and screened for mutants showing differences in pigmentation of immature seeds. The pale banyuls1 (pab1) mutation was found to show reduced levels of anthocyanins in immature seed coat. Here we characterize this mutant at the molecular level.
 , isolated from freshwater fish in Japan
, isolated from freshwater fish in JapanSato, Katsuya; Onodera, Takefumi*; Omoso, Kota*; Takeda-Yano, Kiyoko*; Katayama, Takeshi*; Ono, Yutaka; Narumi, Issey*
Genome Announcements (Internet), 4(1), p.e01631-15_1 - e01631-15_2, 2016/01
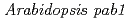 , a mutant with reduced anthocyanins in immature seeds from
, a mutant with reduced anthocyanins in immature seeds from 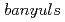 , harbors a mutation in the MATE transporter
, harbors a mutation in the MATE transporter 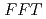
Kitamura, Satoshi; Ono, Yutaka; Narumi, Issey*
Plant Molecular Biology, 90(1-2), p.7 - 18, 2016/01
Times Cited Count:21 Percentile:65.13(Biochemistry & Molecular Biology)Here, we used the Arabidopsis banyuls (ban) mutant, which accumulates anthocyanins, instead of colorless proanthocyanidin precursors, in immature seeds. We mutagenized ban plants and screened for mutants showing differences in pigmentation of immature seeds. The pale banyuls1 (pab1) mutation caused reduced anthocyanin pigmentation in immature seeds compared with ban, but showed normal expression of anthocyanin biosynthetic genes. Map-based cloning showed that two independent pab1 alleles disrupted the MATE-type transporter gene FFT/DTX35. Complementation of pab1 with FFT confirmed that mutation in FFT causes the pab1 phenotype. During development, FFT promoter activity was detected in the seed-coat layers that accumulate flavonoids. Anthocyanins accumulate in the vacuole and FFT fused to GFP mainly localized in the vacuolar membrane.
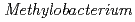 sp. ME121, isolated from soil as a mixed single colony with
sp. ME121, isolated from soil as a mixed single colony with 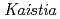 sp. 32K
sp. 32KFujinami, Shun*; Takeda, Kiyoko*; Onodera, Takefumi*; Sato, Katsuya; Shimizu, Tetsu*; Wakabayashi, Yu*; Narumi, Issey*; Nakamura, Akira*; Ito, Masahiro*
Genome Announcements (Internet), 3(5), p.e01005-15_1 - e01005-15_2, 2015/09
Ono, Yutaka; Hase, Yoshihiro; Sato, Katsuya; Nozawa, Shigeki; Narumi, Issey*
Hoshasen To Sangyo, (138), p.17 - 20, 2015/06
no abstracts in English
 exhibiting different anthocyanin accumulation patterns
exhibiting different anthocyanin accumulation patternsKitamura, Satoshi; Ono, Yutaka; Narumi, Issey*
JAEA-Review 2014-050, JAEA Takasaki Annual Report 2013, P. 72, 2015/03
Ion beams have been used for isolation of novel mutants in a number of plant species in our laboratory. Re-mutagenesis by secondary irradiation of ion beams to already-established ion beam-induced mutants would be effective to modify plant characters step-by-step, because ion beams are likely to induce mutations at the limited regions of the genome. Here, we try to adapt the re-mutagenesis by secondary ion beam irradiation in model plant  , for obtaining flavonoid mutants that have not been found so far by standard mutagenesis and screening methods.
, for obtaining flavonoid mutants that have not been found so far by standard mutagenesis and screening methods.
Sato, Katsuya; Ueda, Ryoshiro; Hase, Yoshihiro; Narumi, Issey*; Ono, Yutaka
JAEA-Review 2014-050, JAEA Takasaki Annual Report 2013, P. 117, 2015/03

Ueda, Ryoshiro; Sato, Katsuya; Hayashi, Hidenori*; Narumi, Issey*; Ono, Yutaka
JAEA-Review 2014-050, JAEA Takasaki Annual Report 2013, P. 118, 2015/03
 USDA110 generated by ion-beam irradiation
USDA110 generated by ion-beam irradiationTakeda, Kiyoko*; Sato, Katsuya; Narumi, Issey*; Ono, Yutaka; Otsu, Naoko*; Yokoyama, Tadashi*
JAEA-Review 2014-050, JAEA Takasaki Annual Report 2013, P. 120, 2015/03
 ray-induced mutagenesis
ray-induced mutagenesisSaito, Tsutomu*; Fitriana, Y.*; Sato, Katsuya; Ono, Yutaka; Narumi, Issey*
JAEA-Review 2014-050, JAEA Takasaki Annual Report 2013, P. 121, 2015/03
Nunoshiba, Tatsuo*; Yamauchi, Ayako*; Iwata, Rika*; Sato, Katsuya; Ono, Yutaka; Narumi, Issey*
JAEA-Review 2014-050, JAEA Takasaki Annual Report 2013, P. 124, 2015/03
 (Hypocreales: Clavicipitaceae) mutants induced by ion beams
(Hypocreales: Clavicipitaceae) mutants induced by ion beamsFitriana, Y.*; Shinohara, Shinobu*; Sato, Katsuya; Narumi, Issey*; Saito, Tsutomu*
Applied Entomology and Zoology, 50(1), p.123 - 129, 2015/02
Times Cited Count:0 Percentile:1.75(Chemistry, Physical) L. irradiated by
L. irradiated by  rays
raysKitano, Sayaka*; Miyagi, Atsuko*; Ono, Yutaka; Hase, Yoshihiro; Narumi, Issey*; Yamaguchi, Masatoshi*; Uchimiya, Hirofumi*; Kawai, Maki*
Metabolomics, 11(1), p.134 - 142, 2015/02
Times Cited Count:8 Percentile:25.41(Endocrinology & Metabolism)Kawasaki, Michio*; Kikyo, Shogo*; Nozawa, Shigeki; Akita, Yusuke*; Hase, Yoshihiro; Narumi, Issey*
Nihon Sakumotsu Gakkai Tohoku Shibu Kaiho, (57), p.61 - 62, 2014/12
no abstracts in English
 AV1934, a classic alkaliphile isolated from human feces in 1934
AV1934, a classic alkaliphile isolated from human feces in 1934Attie, O.*; Jayaprakash, A.*; Shah, H.*; Paulsen, I. T.*; Morino, Masato*; Takahashi, Yuka*; Narumi, Issey*; Sachidanandam, R.*; Sato, Katsuya; Ito, Masahiro*; et al.
Genome Announcements (Internet), 2(6), p.e01175-14_1 - e01175-14_2, 2014/11

Adachi, Motoyasu; Hirayama, Hiroshi; Shimizu, Rumi; Sato, Katsuya; Narumi, Issey*; Kuroki, Ryota
Protein Science, 23(10), p.1349 - 1358, 2014/10
Times Cited Count:9 Percentile:25.51(Biochemistry & Molecular Biology)Pleiotropic protein promoting DNA repair A (PprA) is a key protein that facilitates the extreme radioresistance of  . To clarify the role of PprA in the radioresistance mechanism, the interaction between recombinant PprA expressed in Escherichia coli with several double-stranded DNAs was investigated. In a gel-shift assay, the band shift of supercoiled pUC19 DNA caused by the binding of PprA showed a bimodal distribution, which was promoted by the addition of 1 mM Mg, Ca, or Sr ions. The dissociation constant of the PprA-supercoiled pUC19 DNA complex, calculated from the relative portions of shifted bands, was 0.6
. To clarify the role of PprA in the radioresistance mechanism, the interaction between recombinant PprA expressed in Escherichia coli with several double-stranded DNAs was investigated. In a gel-shift assay, the band shift of supercoiled pUC19 DNA caused by the binding of PprA showed a bimodal distribution, which was promoted by the addition of 1 mM Mg, Ca, or Sr ions. The dissociation constant of the PprA-supercoiled pUC19 DNA complex, calculated from the relative portions of shifted bands, was 0.6 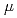 M with a Hill coefficient of 3.3 in the presence of 1 mM Mg acetate. This indicates that at least 281 PprA molecules are required to saturate a supercoiled pUC19 DNA, which is consistent with the number of bound PprA molecules estimated by the UV absorption of the PprA-pUC19 complex purified by gel filtration. This saturation also suggests linear polymerization of PprA along the dsDNA. On the other hand, the bands of linear dsDNA and nicked circular dsDNA that eventually formed PprA complexes did not saturate, but created larger molecular complexes when the PprA concentration was greater than 1.3
M with a Hill coefficient of 3.3 in the presence of 1 mM Mg acetate. This indicates that at least 281 PprA molecules are required to saturate a supercoiled pUC19 DNA, which is consistent with the number of bound PprA molecules estimated by the UV absorption of the PprA-pUC19 complex purified by gel filtration. This saturation also suggests linear polymerization of PprA along the dsDNA. On the other hand, the bands of linear dsDNA and nicked circular dsDNA that eventually formed PprA complexes did not saturate, but created larger molecular complexes when the PprA concentration was greater than 1.3 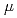 M. This result implies that DNA-bound PprA aids association of the termini of damaged DNAs, which is regulated by the concentration of PprA.
M. This result implies that DNA-bound PprA aids association of the termini of damaged DNAs, which is regulated by the concentration of PprA.
 sp. strain TCA20, isolated from a hot spring containing a high concentration of calcium ions
sp. strain TCA20, isolated from a hot spring containing a high concentration of calcium ionsFujinami, Shun*; Takeda, Kiyoko*; Onodera, Takefumi*; Sato, Katsuya; Sano, Motohiko*; Takahashi, Yuka*; Narumi, Issey*; Ito, Masahiro*
Genome Announcements (Internet), 2(5), p.e00866-14_1 - e00866-14_2, 2014/09