Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
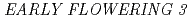 and
and 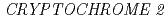 genes delays flowering under continuous light but accelerates it under long days and short days; An Important role for
genes delays flowering under continuous light but accelerates it under long days and short days; An Important role for  CRY2 to accelerate flowering time in continuous light
CRY2 to accelerate flowering time in continuous lightNefissi, R.*; Natsui, Yu*; Miyata, Kana*; Oda, Atsushi*; Hase, Yoshihiro; Nakagawa, Mayu*; Ghorbel, A.*; Mizoguchi, Tsuyoshi*
Journal of Experimental Botany, 62(8), p.2731 - 2744, 2011/02
Times Cited Count:18 Percentile:49.43(Plant Sciences)no abstracts in English
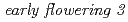 in
in 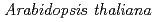
Natsui, Yu*; Nefissi, R.*; Miyata, Kana*; Oda, Atsushi*; Hase, Yoshihiro; Nakagawa, Mayu*; Mizoguchi, Tsuyoshi*
Plant Biotechnology, 27(5), p.463 - 468, 2010/12
Times Cited Count:4 Percentile:15.29(Biotechnology & Applied Microbiology)EARLY FLOWERING 3 (ELF3) is a circadian clock protein with a major role in maintaining circadian rhythms in plants. In this work, 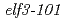 was mutagenized by EMS in plants of the Lansberg
was mutagenized by EMS in plants of the Lansberg  background to isolate suppressors of
background to isolate suppressors of 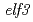 and to understand the molecular mechanisms of flowering time controlled by ELF3. Two suppressors,
and to understand the molecular mechanisms of flowering time controlled by ELF3. Two suppressors, 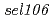 obtained from this screen and
obtained from this screen and 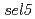 from a precious study, were chosen for further analysis. Genetic mapping, gene expression analysis, and sequencing identified
from a precious study, were chosen for further analysis. Genetic mapping, gene expression analysis, and sequencing identified 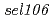 and
and 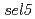 as new alleles of
as new alleles of 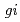 and
and 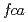 , respectively. Genetic interactions between
, respectively. Genetic interactions between 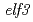 and
and 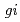 and between
and between 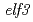 and
and 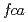 in the control of the floral activator
in the control of the floral activator 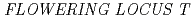 were also investigated. Six suppressor of
were also investigated. Six suppressor of 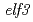 were classified at least into four subgroups based on the expression of such floral regulators as GI, CO, FT, SVP, and FLC, and on their flowering times under LL, LD, and SD. This classification scheme is useful for the characterization of unidentified suppressor mutations.
were classified at least into four subgroups based on the expression of such floral regulators as GI, CO, FT, SVP, and FLC, and on their flowering times under LL, LD, and SD. This classification scheme is useful for the characterization of unidentified suppressor mutations.
Natsui, Yu*; Nefissi, R.*; Miyata, Kana*; Oda, Atsushi*; Hase, Yoshihiro; Nakagawa, Mayu; Mizoguchi, Tsuyoshi*
no journal, ,
EARLY FLOWERING 3 (ELF3) plays key roles in the control of light response, organ elongation, flowering time and circadian rhythms in Arabidopsis. ELF3 does not show significant homology to any other proteins in the public databases. Therefore biochemical function of the ELF3 has not been well understood. To understand molecular mechanisms underlying the a variety of phenotypes in the elf3 mutants, suppressors of elf3-1 were screened for, using the elf3-1 seeds mutagenized with heavy ion beams. Here we demonstrate that in one of the suppressor lines, S#20, early flowering, small cotyledon and pale green phenotypes of elf3-1 were suppressed, whereas long hypocotyl phenotype was enhanced. The recessive suppressor mutation was named suppressor of elf3 20 (sel20) and mapped to the upper side of Ch1. We have found 21-bp deletion in the FHA/CRY2 gene in the sel20. We will discuss possible roles of CRY2 in the multiple functions of ELF3.