Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Chiba, Kaori*; Matsui, Takuro*; Chatake, Toshiyuki*; Ohara, Takashi; Tanaka, Ichiro*; Yutani, Katsuhide*; Niimura, Nobuo*
Protein Science, 32(10), p.e4765_1 - e4765_13, 2023/10
Times Cited Count:0 Percentile:0.00(Biochemistry & Molecular Biology)Yano, Naomine*; Yamada, Taro*; Hosoya, Takaaki*; Ohara, Takashi; Tanaka, Ichiro*; Niimura, Nobuo*; Kusaka, Katsuhiro*
Acta Crystallographica Section D; Structural Biology (Internet), 74(11), p.1041 - 1052, 2018/11
Times Cited Count:16 Percentile:73.72(Biochemical Research Methods)Tashiro, Koji*; Kusaka, Katsuhiro*; Hosoya, Takaaki*; Ohara, Takashi; Hanesaka, Makoto*; Yoshizawa, Yoshinori*; Yamamoto, Hiroko*; Niimura, Nobuo*; Tanaka, Ichiro*; Kurihara, Kazuo*; et al.
Macromolecules, 51(11), p.3911 - 3922, 2018/06
Times Cited Count:6 Percentile:18.40(Polymer Science)Unno, Masayoshi*; Ishikawa, Kumiko*; Kusaka, Katsuhiro*; Tamada, Taro; Hagiwara, Yoshinori*; Sugishima, Masakazu*; Wada, Kei*; Yamada, Taro*; Tomoyori, Katsuaki; Hosoya, Takaaki*; et al.
Journal of the American Chemical Society, 137(16), p.5452 - 5460, 2015/04
Times Cited Count:29 Percentile:62.63(Chemistry, Multidisciplinary)Phycocyanobilin, a light-harvesting and photoreceptor pigment in higher plants, algae, and cyanobacteria, is synthesized from biliverdin IX (BV) by phycocyanobilin:ferredoxin oxidoreductase (PcyA) via two steps of two-proton-coupled two-electron reduction. We determined the neutron structure of PcyA from cyanobacteria complexed with BV, revealing the exact location of the hydrogen atoms involved in catalysis. Notably, approximately half of the BV bound to PcyA was BVH
(BV) by phycocyanobilin:ferredoxin oxidoreductase (PcyA) via two steps of two-proton-coupled two-electron reduction. We determined the neutron structure of PcyA from cyanobacteria complexed with BV, revealing the exact location of the hydrogen atoms involved in catalysis. Notably, approximately half of the BV bound to PcyA was BVH , a state in which all four pyrrole nitrogen atoms were protonated. The protonation states of BV complemented the protonation of adjacent Asp105. The "axial "water molecule that interacts with the neutral pyrrole nitrogen of the A-ring was identified. His88 N
, a state in which all four pyrrole nitrogen atoms were protonated. The protonation states of BV complemented the protonation of adjacent Asp105. The "axial "water molecule that interacts with the neutral pyrrole nitrogen of the A-ring was identified. His88 N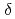 was protonated to form a hydrogen bond with the lactam O atom of the BV A-ring. His88 and His74 were linked by hydrogen bonds via H
was protonated to form a hydrogen bond with the lactam O atom of the BV A-ring. His88 and His74 were linked by hydrogen bonds via H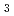 O
O . These results imply that Asp105, His88, and the axial water molecule contribute to proton transfer during PcyA catalysis.
. These results imply that Asp105, His88, and the axial water molecule contribute to proton transfer during PcyA catalysis.
Tashiro, Koji*; Hanesaka, Makoto*; Yamamoto, Hiroko*; Wasanasuk, K.*; Jayaratri, P.*; Yoshizawa, Yoshinori*; Tanaka, Ichiro*; Niimura, Nobuo*; Kusaka, Katsuhiro*; Hosoya, Takaaki*; et al.
Kobunshi Rombunshu, 71(11), p.508 - 526, 2014/11
Times Cited Count:6 Percentile:20.53(Polymer Science)The crystal structure analysis of various polymer substances has been reviewed on the basis of wide-angle high-energy X-ray and neutron diffraction data. The progress in structural analytical techniques of polymer crystals have been reviewed at first. The structural models proposed so far were reinvestigated and new models have been proposed for various kinds of polymer crystals including polyethylene, poly(vinyl alcohol), poly(lactic acid) and its stereocomplex etc. The hydrogen atomic positions were also clarified by the quantitative analysis of wide-angle neutron diffraction data, from which the physical properties of polymer crystals have been evaluated theoretically. The bonded electron density distribution has been estimated for a polydiacetylene single crystal on the basis of the so-called X-N method or by the combination of structural information derived from X-ray and neutron diffraction data analysis. Some comments have been added about future developments in the field of structure-property relationship determination.
Kusaka, Katsuhiro*; Hosoya, Takaaki*; Yamada, Taro*; Tomoyori, Katsuaki; Ohara, Takashi; Katagiri, Masaki*; Kurihara, Kazuo; Tanaka, Ichiro*; Niimura, Nobuo*
Journal of Synchrotron Radiation, 20(6), p.994 - 998, 2013/11
Times Cited Count:39 Percentile:85.54(Instruments & Instrumentation)The IBARAKI biological crystal diffractometer, iBIX, is a high-performance time-of-flight neutron single-crystal diffractometer for elucidating mainly the hydrogen, protonation and hydration structures of biological macromolecules in various life processes. Since the end of 2008, iBIX has been available to user's experiments supported by Ibaraki University. Since August 2012, an upgrade of the 14-existing detectors has begun and 16 new detectors have been installed for iBIX. The total measurement efficiency of the present diffractometer has been impoved by one order of magnitude from the previous one with the increasing of accelerator power. In December 2012, commissioning of the new detectors was successful, and collection of the diffraction dataset of ribonucrease A as a standard protein was attempted in order to estimate the performance of the upgraded iBIX in comparison with previous results. The resolution of diffraction data, equivalence among intensities of symmetry-related reflections and reliability of the refined structure have been improved dramatically. iBIX is expected to be one of the highest-performance neutron single-crystal diffractometers for biological macromolecules in the world.
Yokoyama, Takeshi*; Mizuguchi, Mineyuki*; Nabeshima, Yuko*; Kusaka, Katsuhiro*; Yamada, Taro*; Hosoya, Takaaki*; Ohara, Takashi; Kurihara, Kazuo; Tanaka, Ichiro*; Niimura, Nobuo*
Journal of Synchrotron Radiation, 20(6), p.834 - 837, 2013/11
Times Cited Count:7 Percentile:36.12(Instruments & Instrumentation)Transthyretin (TTR) is a tetrameric protein. TTR misfolding and aggregation are associated with human amyloid diseases. Dissociation of the TTR tetramer is believed to be the rate-limiting step in the amyloid fibril formation cascade. Low pH is known to promote dissociation into monomer and the formation of amyloid fibrils. In order to reveal the molecular mechanisms underlying pH sensitivity and structural stabilities of TTR, neutron diffraction studies were conducted using the IBARAKI Biological Crystal Diffractometer with the time-of-flight method. Crystals for the neutron diffraction experiments were grown up to 2.5 mm for four months. The neutron crystal structure solved at 2.0
for four months. The neutron crystal structure solved at 2.0  revealed the protonation states of His88 and the detailed hydrogen-bond network depending on the protonation states of His88. This hydrogen-bond network is involved in monomer-monomer and dimer-dimer interactions, suggesting that the double protonation of His88 by acidification breaks the hydrogen-bond network and causes the destabilization of the TTR tetramer. Structural comparison with the X-ray crystal structure at acidic pH identified the three amino acid residues responsible for the pH sensitivity of TTR. Our neutron model provides insights into the molecular stability related to amyloidosis.
revealed the protonation states of His88 and the detailed hydrogen-bond network depending on the protonation states of His88. This hydrogen-bond network is involved in monomer-monomer and dimer-dimer interactions, suggesting that the double protonation of His88 by acidification breaks the hydrogen-bond network and causes the destabilization of the TTR tetramer. Structural comparison with the X-ray crystal structure at acidic pH identified the three amino acid residues responsible for the pH sensitivity of TTR. Our neutron model provides insights into the molecular stability related to amyloidosis.
 -thrombin-bivalirubin complex at pD 5.0; Protonation states and hydration structure of the enzyme-product complex
-thrombin-bivalirubin complex at pD 5.0; Protonation states and hydration structure of the enzyme-product complexYamada, Taro*; Kurihara, Kazuo; Onishi, Yuki*; Tamada, Taro; Tomoyori, Katsuaki; Masumi, Kenji*; Tanaka, Ichiro*; Kuroki, Ryota; Niimura, Nobuo*
Biochimica et Biophysica Acta; Proteins and Proteomics, 1834(8), p.1532 - 1538, 2013/08
Times Cited Count:20 Percentile:52.35(Biochemistry & Molecular Biology)The protonation states and hydration structures of the  -thrombin-bivalirubin complex were studied by joint XN refinement of the single crystal X-ray and neutron diffraction data at resolutions of 1.6 and 2.8
-thrombin-bivalirubin complex were studied by joint XN refinement of the single crystal X-ray and neutron diffraction data at resolutions of 1.6 and 2.8  , respectively. The atomic distances were estimated by carrying out X-ray crystallographic analysis at 1.25
, respectively. The atomic distances were estimated by carrying out X-ray crystallographic analysis at 1.25  resolution. The complex represents a model of the enzyme-product (EP) complex of
resolution. The complex represents a model of the enzyme-product (EP) complex of  -thrombin. The neutron scattering length maps around the active site suggest that the side chain of H57/H was deuterated. The joint XN refinement showed that occupancies for D
-thrombin. The neutron scattering length maps around the active site suggest that the side chain of H57/H was deuterated. The joint XN refinement showed that occupancies for D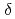 1 and D
1 and D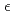 2 of H57/H were 1.0 and 0.7, respectively. However, no significant neutron scattering length density was observed around the hydroxyl oxygen O
2 of H57/H were 1.0 and 0.7, respectively. However, no significant neutron scattering length density was observed around the hydroxyl oxygen O of S195/H, which was close to the carboxylic carbon atom of dFPR-COOH. These observations suggest that the O
of S195/H, which was close to the carboxylic carbon atom of dFPR-COOH. These observations suggest that the O atom of S195/H is deprotonated and maintains its nucleophilicity in the EP complex. In addition to the active site, the hydration structures of the S1 subsite and the Exosite I, which are involved in the recognition of bivalirudin, are presented.
atom of S195/H is deprotonated and maintains its nucleophilicity in the EP complex. In addition to the active site, the hydration structures of the S1 subsite and the Exosite I, which are involved in the recognition of bivalirudin, are presented.
 protease I at pD 8.0; Protonation states and hydration structure in the free-form
protease I at pD 8.0; Protonation states and hydration structure in the free-formOnishi, Yuki*; Yamada, Taro*; Kurihara, Kazuo; Tanaka, Ichiro*; Sakiyama, Fumio*; Masaki, Takeharu*; Niimura, Nobuo*
Biochimica et Biophysica Acta; Proteins and Proteomics, 1834(8), p.1642 - 1647, 2013/08
Times Cited Count:8 Percentile:22.43(Biochemistry & Molecular Biology)The structure of the free-form of  protease I (API) at pD 8.0 was refined by simultaneous use of single crystal X-ray and neutron diffraction data sets to investigate the protonation states of key catalytic residues of the serine protease. Occupancy refinement of the catalytic triad in the active site of API free-form showed that ca. 30% of the imidazole ring of H57 and ca. 70% of the hydroxyl group of S194 were deuterated. This observation indicates that a major fraction of S194 is protonated in the absence of a substrate. The protonation state of the catalytic triad in API was compared with the bovine
protease I (API) at pD 8.0 was refined by simultaneous use of single crystal X-ray and neutron diffraction data sets to investigate the protonation states of key catalytic residues of the serine protease. Occupancy refinement of the catalytic triad in the active site of API free-form showed that ca. 30% of the imidazole ring of H57 and ca. 70% of the hydroxyl group of S194 were deuterated. This observation indicates that a major fraction of S194 is protonated in the absence of a substrate. The protonation state of the catalytic triad in API was compared with the bovine  -trypsin-BPTI complex. The comparison led to the hypothesis that close contact of a substrate with S194 could lower the acidity of its hydroxyl group, thereby allowing H57 to extract the hydrogen from the hydroxyl group of S194. H210, which is a residue specific to API, does not form a hydrogen bond with the catalytic triad residue D113. Instead, H210 forms a hydrogen bond network with S176, H177 and a water molecule. The close proximity of the bulky, hydrophobic residue W169 may protect this hydrogen bond network, and this protection may stabilize the function of API over a wide pH range.
-trypsin-BPTI complex. The comparison led to the hypothesis that close contact of a substrate with S194 could lower the acidity of its hydroxyl group, thereby allowing H57 to extract the hydrogen from the hydroxyl group of S194. H210, which is a residue specific to API, does not form a hydrogen bond with the catalytic triad residue D113. Instead, H210 forms a hydrogen bond network with S176, H177 and a water molecule. The close proximity of the bulky, hydrophobic residue W169 may protect this hydrogen bond network, and this protection may stabilize the function of API over a wide pH range.
Yokoyama, Takeshi*; Mizuguchi, Mineyuki*; Nabeshima, Yuko*; Kusaka, Katsuhiro*; Yamada, Taro*; Hosoya, Takaaki*; Ohara, Takashi*; Kurihara, Kazuo; Tomoyori, Katsuaki*; Tanaka, Ichiro*; et al.
Journal of Structural Biology, 177(2), p.283 - 290, 2012/02
Times Cited Count:49 Percentile:82.30(Biochemistry & Molecular Biology)Kawamura, Kenji*; Yamada, Taro*; Kurihara, Kazuo; Tamada, Taro; Kuroki, Ryota; Tanaka, Ichiro*; Takahashi, Haruyuki*; Niimura, Nobuo*
Acta Crystallographica Section D, 67(2), p.140 - 148, 2011/02
Times Cited Count:30 Percentile:88.81(Biochemical Research Methods)Tanaka, Ichiro*; Kusaka, Katsuhiro*; Hosoya, Takaaki*; Niimura, Nobuo*; Ohara, Takashi*; Kurihara, Kazuo; Yamada, Taro*; Onishi, Yuki*; Tomoyori, Katsuaki*; Yokoyama, Takeshi*
Acta Crystallographica Section D, 66(11), p.1194 - 1197, 2010/11
Times Cited Count:53 Percentile:94.69(Biochemical Research Methods)The IBARAKI Biological Crystal Diffractometer (iBIX), a new diffractometer for protein crystallography at the next-generation neutron source at J-PARC (Japan Proton Accelerator Research Complex), has been constructed and has been operational since December 2008. Preliminary structure analyses of organic crystals showed that iBIX has high performance even at 120 kW operation and the first full data set is being collected from a protein crystal.
Tanaka, Ichiro*; Kusaka, Katsuhiro*; Hosoya, Takaaki*; Ohara, Takashi*; Kurihara, Kazuo; Niimura, Nobuo*
Yakugaku Zasshi, 130(5), p.665 - 670, 2010/05
Times Cited Count:0 Percentile:0.00(Pharmacology & Pharmacy)Ibaraki Prefectural Government together with Ibaraki University and Japan Atomic Energy Agency (JAEA) has almost finished constructing a time-of-flight (TOF) neutron diffractometer for biological macromolecules for industrial use at J-PARC, IBARAKI Biological Crystal Diffractometer (iBIX). Since 2009, Ibaraki University has been asked to operate this machine in order for users to do experiments by Ibaraki Prefecture. The diffractometer is designed to cover sample crystals which have their cell edges up to around 150  . It is expected to measure more than 100 samples per year if they have 2 mm
. It is expected to measure more than 100 samples per year if they have 2 mm in crystal volume, and to measure even around 0.1 mm
in crystal volume, and to measure even around 0.1 mm in crystal volume of biological samples. The efficiency of iBIX is also expected about 100 times larger than those of the present high performance diffractometers at JRR-3 in JAEA when 1MW power realizes in J-PARC. Since December 2008, iBIX has been open to users and several proteins and organic compounds were tested under 20 kW proton power of J-PARC. It was found that one of their proteins was diffracted up to 1.4
in crystal volume of biological samples. The efficiency of iBIX is also expected about 100 times larger than those of the present high performance diffractometers at JRR-3 in JAEA when 1MW power realizes in J-PARC. Since December 2008, iBIX has been open to users and several proteins and organic compounds were tested under 20 kW proton power of J-PARC. It was found that one of their proteins was diffracted up to 1.4  in d-spacing, which was nearly comparable resolution to that of BIX-3 in JRR-3 when used the same crystal as at iBIX for reasonable exposure time. In May 2009, 14 detector units were set up. By the end of fiscal year 2009, the basic part of data reduction software will be finished and an equipment blowing low temperature gas to the sample will be installed with the cooperation of JAEA.
in d-spacing, which was nearly comparable resolution to that of BIX-3 in JRR-3 when used the same crystal as at iBIX for reasonable exposure time. In May 2009, 14 detector units were set up. By the end of fiscal year 2009, the basic part of data reduction software will be finished and an equipment blowing low temperature gas to the sample will be installed with the cooperation of JAEA.
 resolution
resolutionYagi, Daichi*; Yamada, Taro*; Kurihara, Kazuo; Onishi, Yuki*; Yamashita, Masahiro*; Tamada, Taro; Tanaka, Ichiro*; Kuroki, Ryota; Niimura, Nobuo*
Acta Crystallographica Section D, 65(9), p.892 - 899, 2009/09
Times Cited Count:17 Percentile:79.69(Biochemical Research Methods)A neutron crystallographic analysis of phosphate-free bovine pancreatic RNase A has been carried out at 1.7  resolution using the BIX-4 single-crystal diffractometer at the JRR-3 reactor of the Japan Atomic Energy Agency. The high resolution structural model allowed us to determine that His12 acts mainly as a general base in the catalytic process of RNase A. Numerous other distinctive structural features such as the hydrogen positions of methyl groups, hydroxyl groups, prolines, asparagines and glutamines were also determined at 1.7
resolution using the BIX-4 single-crystal diffractometer at the JRR-3 reactor of the Japan Atomic Energy Agency. The high resolution structural model allowed us to determine that His12 acts mainly as a general base in the catalytic process of RNase A. Numerous other distinctive structural features such as the hydrogen positions of methyl groups, hydroxyl groups, prolines, asparagines and glutamines were also determined at 1.7  resolution. The protonation and deprotonation states of all of the charged amino-acid residues allowed us to provide a definitive description of the hydrogen-bonding network around the active site and the H atoms of the key His48 residue. Differences in hydrogen-bond strengths for the
resolution. The protonation and deprotonation states of all of the charged amino-acid residues allowed us to provide a definitive description of the hydrogen-bonding network around the active site and the H atoms of the key His48 residue. Differences in hydrogen-bond strengths for the  -helices and
-helices and  -sheets were inferred from determination of the hydrogen-bond lengths and the H/D-exchange ratios of the backbone amide H atoms. The correlation between the B factors and hydrogen-bond lengths of the hydration water molecules was also determined.
-sheets were inferred from determination of the hydrogen-bond lengths and the H/D-exchange ratios of the backbone amide H atoms. The correlation between the B factors and hydrogen-bond lengths of the hydration water molecules was also determined.
Yamaguchi, Shigeo*; Kamikubo, Hironari*; Kurihara, Kazuo; Kuroki, Ryota; Niimura, Nobuo*; Shimizu, Nobutaka*; Yamazaki, Yoichi*; Kataoka, Mikio*
Proceedings of the National Academy of Sciences of the United States of America, 106(2), p.440 - 444, 2009/01
Times Cited Count:163 Percentile:94.50(Multidisciplinary Sciences)Fujii, Yasuhiko; Arai, Masatoshi; Kadono, Ryosuke*; Kanaya, Toshiji*; Kamiyama, Takashi*; Niimura, Nobuo*; Nojiri, Hiroyuki*; Noda, Yukio*; Yagi, Takehiko*; Yamada, Kazuyoshi*
Kotai Butsuri, 43(7), p.441 - 450, 2008/07
no abstracts in English
Ishikawa, Takuya*; Chatake, Toshiyuki*; Onishi, Yuki*; Tanaka, Ichiro*; Kurihara, Kazuo; Kuroki, Ryota; Niimura, Nobuo*
Chemical Physics, 345(2-3), p.152 - 158, 2008/04
Times Cited Count:14 Percentile:43.46(Chemistry, Physical)Yamasaki, Chisato*; Murakami, Katsuhiko*; Fujii, Yasuyuki*; Sato, Yoshiharu*; Harada, Erimi*; Takeda, Junichi*; Taniya, Takayuki*; Sakate, Ryuichi*; Kikugawa, Shingo*; Shimada, Makoto*; et al.
Nucleic Acids Research, 36(Database), p.D793 - D799, 2008/01
Times Cited Count:52 Percentile:70.47(Biochemistry & Molecular Biology)Here we report the new features and improvements in our latest release of the H-Invitational Database, a comprehensive annotation resource for human genes and transcripts. H-InvDB, originally developed as an integrated database of the human transcriptome based on extensive annotation of large sets of fulllength cDNA (FLcDNA) clones, now provides annotation for 120 558 human mRNAs extracted from the International Nucleotide Sequence Databases (INSD), in addition to 54 978 human FLcDNAs, in the latest release H-InvDB. We mapped those human transcripts onto the human genome sequences (NCBI build 36.1) and determined 34 699 human gene clusters, which could define 34 057 protein-coding and 642 non-protein-coding loci; 858 transcribed loci overlapped with predicted pseudogenes.
Chatake, Toshiyuki*; Shibayama, Naoya*; Park, S. Y.*; Kurihara, Kazuo; Tamada, Taro; Tanaka, Ichiro*; Niimura, Nobuo*; Kuroki, Ryota; Morimoto, Yukio*
Journal of the American Chemical Society, 129(48), p.14840 - 14841, 2007/12
Times Cited Count:27 Percentile:60.46(Chemistry, Multidisciplinary)Tashiro, Koji*; Hanesaka, Makoto*; Ohara, Takashi; Ozeki, Tomoji*; Kitano, Toshiaki*; Nishu, Takashi*; Kurihara, Kazuo; Tamada, Taro; Kuroki, Ryota; Fujiwara, Satoru; et al.
Polymer Journal, 39(12), p.1253 - 1273, 2007/12
Times Cited Count:21 Percentile:53.40(Polymer Science)no abstracts in English