Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
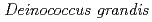 , isolated from freshwater fish in Japan
, isolated from freshwater fish in JapanSato, Katsuya; Onodera, Takefumi*; Omoso, Kota*; Takeda-Yano, Kiyoko*; Katayama, Takeshi*; Ono, Yutaka; Narumi, Issey*
Genome Announcements (Internet), 4(1), p.e01631-15_1 - e01631-15_2, 2016/01
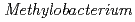 sp. ME121, isolated from soil as a mixed single colony with
sp. ME121, isolated from soil as a mixed single colony with 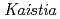 sp. 32K
sp. 32KFujinami, Shun*; Takeda, Kiyoko*; Onodera, Takefumi*; Sato, Katsuya; Shimizu, Tetsu*; Wakabayashi, Yu*; Narumi, Issey*; Nakamura, Akira*; Ito, Masahiro*
Genome Announcements (Internet), 3(5), p.e01005-15_1 - e01005-15_2, 2015/09
 sp. strain TCA20, isolated from a hot spring containing a high concentration of calcium ions
sp. strain TCA20, isolated from a hot spring containing a high concentration of calcium ionsFujinami, Shun*; Takeda, Kiyoko*; Onodera, Takefumi*; Sato, Katsuya; Sano, Motohiko*; Takahashi, Yuka*; Narumi, Issey*; Ito, Masahiro*
Genome Announcements (Internet), 2(5), p.e00866-14_1 - e00866-14_2, 2014/09
 sp. strain TS-2, isolated from a jumping spider
sp. strain TS-2, isolated from a jumping spiderFujinami, Shun*; Takeda, Kiyoko*; Onodera, Takefumi*; Sato, Katsuya; Sano, Motohiko*; Narumi, Issey*; Ito, Masahiro*
Genome Announcements (Internet), 2(3), p.e00458-14_1 - e00458-14_2, 2014/05

Sato, Katsuya; Onodera, Takefumi*; Takeda, Kiyoko; Narumi, Issey*
JAEA-Review 2013-059, JAEA Takasaki Annual Report 2012, P. 110, 2014/03
 and
and 
Onodera, Takefumi*; Sato, Katsuya; Ota, Toshihiro*; Narumi, Issey*
JAEA-Review 2013-059, JAEA Takasaki Annual Report 2012, P. 111, 2014/03
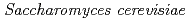
Nunoshiba, Tatsuo*; Sakata, Yayoi*; Yamauchi, Ayako*; Sato, Katsuya; Onodera, Takefumi*; Narumi, Issey*
JAEA-Review 2013-059, JAEA Takasaki Annual Report 2012, P. 118, 2014/03
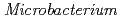 sp. strain TS-1
sp. strain TS-1Fujinami, Shun*; Takeda, Kiyoko; Onodera, Takefumi*; Sato, Katsuya; Sano, Motohiko*; Narumi, Issey*; Ito, Masahiro*
Genome Announcements (Internet), 1(6), P. e01043-13, 2013/12
 YgjD and YeaZ are involved in the repair of DNA cross-links
YgjD and YeaZ are involved in the repair of DNA cross-linksOnodera, Takefumi; Sato, Katsuya; Ota, Toshihiro*; Narumi, Issei
Extremophiles, 17(1), p.171 - 179, 2013/01
Times Cited Count:10 Percentile:26.85(Biochemistry & Molecular Biology)
Sato, Katsuya; Tejima, Kohei*; Onodera, Takefumi; Narumi, Issei
JAEA-Review 2012-046, JAEA Takasaki Annual Report 2011, P. 103, 2013/01
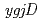 and
and 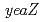 orthologs, in DNA repair of
orthologs, in DNA repair of 
Onodera, Takefumi; Sato, Katsuya; Ota, Toshihiro*; Narumi, Issei
JAEA-Review 2012-046, JAEA Takasaki Annual Report 2011, P. 104, 2013/01
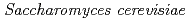
Nunoshiba, Tatsuo*; Hirasawa, Yukei*; Yamauchi, Ayako*; Sato, Katsuya; Tejima, Kohei*; Onodera, Takefumi; Narumi, Issei
JAEA-Review 2012-046, JAEA Takasaki Annual Report 2011, P. 111, 2013/01
 RecFOR proteins in homologous recombination
RecFOR proteins in homologous recombinationSato, Katsuya; Kikuchi, Masahiro; Ishaque, A. M.*; Oba, Hirofumi*; Yamada, Mitsugu; Tejima, Kohei; Onodera, Takefumi; Narumi, Issei
DNA Repair, 11(4), p.410 - 418, 2012/04
Times Cited Count:24 Percentile:59.83(Genetics & Heredity)In an effort to gain insights into the role of 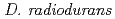 RecFOR proteins in homologous recombination, we generated
RecFOR proteins in homologous recombination, we generated 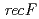 ,
, 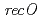 and
and 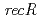 disruptant strains and characterized the disruption effects. Disruption of
disruptant strains and characterized the disruption effects. Disruption of 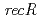 resulted in severe reduction of the transformation efficiency. On the other hand, the
resulted in severe reduction of the transformation efficiency. On the other hand, the 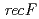 disruptant strain was the most sensitive phenotype to
disruptant strain was the most sensitive phenotype to  rays, UV irradiation and mitomycin C among the three disruptants. In the
rays, UV irradiation and mitomycin C among the three disruptants. In the 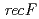 disruptant strain, the intracellular level of the LexA1 protein did not decrease following
disruptant strain, the intracellular level of the LexA1 protein did not decrease following  irradiation. These results demonstrate that the RecF protein plays a crucial role in the homologous recombination repair process by facilitating RecA activation. Thus, the RecF and RecR proteins are involved in the RecA activation and the stability of incoming DNA, respectively, during RecA-mediated homologous recombination processes that initiated the ESDSA pathway in
irradiation. These results demonstrate that the RecF protein plays a crucial role in the homologous recombination repair process by facilitating RecA activation. Thus, the RecF and RecR proteins are involved in the RecA activation and the stability of incoming DNA, respectively, during RecA-mediated homologous recombination processes that initiated the ESDSA pathway in 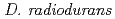 .
.

Sato, Katsuya; Tejima, Kohei; Onodera, Takefumi; Narumi, Issei
JAEA-Review 2011-043, JAEA Takasaki Annual Report 2010, P. 106, 2012/01
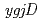 and
and 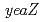 orthologs, in DNA repair of
orthologs, in DNA repair of 
Onodera, Takefumi; Sato, Katsuya; Narumi, Issei; Ota, Toshihiro*
JAEA-Review 2011-043, JAEA Takasaki Annual Report 2010, P. 107, 2012/01
Onodera, Takefumi; Narumi, Issei
Nihon Biseibutsu Seitai Gakkai-Shi, 26(2), p.75 - 77, 2011/09
no abstracts in English
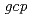 gene, an unknown-essential gene common to living organisms
gene, an unknown-essential gene common to living organismsKitahara, Kazumasa*; Onodera, Takefumi; Hoshino, Takayuki*; Narumi, Issei; Nakamura, Akira*
no journal, ,
no abstracts in English
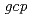 gene, an unknown-essential gene common to living organisms
gene, an unknown-essential gene common to living organismsNakamura, Akira*; Kitahara, Kazumasa*; Onodera, Takefumi; Hoshino, Takayuki*; Narumi, Issei
no journal, ,
no abstracts in English

Sato, Katsuya; Kikuchi, Masahiro; Oba, Hirofumi*; Yamada, Mitsugu; Tejima, Kohei; Onodera, Takefumi; Narumi, Issei
no journal, ,
In an effort to gain an insight into the role of the RecFOR in DNA repair through homologous recombination in  , we generated a RecFOR disruptant strains and characterized the disruption effects. The RecF disruptant strain exhibited sensitivity to DNA-damaging agents, suggesting that RecF has a crucial role in DNA repair. This result indicates that RecF takes an important role in the RecA activation in the homologous recombination process with DNA damage. While, the recombination assay revealed that RecR is essential for the process of homologous intermolecular recombination, suggesting that RecR is important factor in the homologous recombination process without DNA damage. The importance of RecF and RecR in the homologous recombination processes is reversed due to the existence of DNA damage. Therefore,
, we generated a RecFOR disruptant strains and characterized the disruption effects. The RecF disruptant strain exhibited sensitivity to DNA-damaging agents, suggesting that RecF has a crucial role in DNA repair. This result indicates that RecF takes an important role in the RecA activation in the homologous recombination process with DNA damage. While, the recombination assay revealed that RecR is essential for the process of homologous intermolecular recombination, suggesting that RecR is important factor in the homologous recombination process without DNA damage. The importance of RecF and RecR in the homologous recombination processes is reversed due to the existence of DNA damage. Therefore, 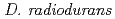 will use the different homologous recombination processes through the RecFOR or RecOR, respectively.
will use the different homologous recombination processes through the RecFOR or RecOR, respectively.

Onodera, Takefumi; Kitahara, Kazumasa*; Nakamura, Akira*; Sato, Katsuya; Ota, Toshihiro*; Narumi, Issei
no journal, ,
 -sialoglycoprotein endopeptidase (Gcp) gene has been annotated with, such as
-sialoglycoprotein endopeptidase (Gcp) gene has been annotated with, such as 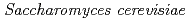 and
and 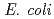 are classified as essential genes. However, in
are classified as essential genes. However, in 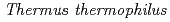 and
and 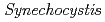 sp. PCC6803, it is not essential. As its function,
sp. PCC6803, it is not essential. As its function, 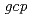 ortholog (
ortholog (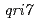 ) mutants in
) mutants in 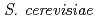 mitochondrial DNA are the loss of genomic DNA. In
mitochondrial DNA are the loss of genomic DNA. In 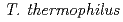 ,
, 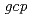 ortholog disruptant was reported that it growth was suppressed under high salt concentration. Mutual relationships between organisms on the role of gcp ortholog has not been found. The study, we performed a genetic analysis of functional roles in
ortholog disruptant was reported that it growth was suppressed under high salt concentration. Mutual relationships between organisms on the role of gcp ortholog has not been found. The study, we performed a genetic analysis of functional roles in 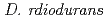 Gcp ortholog encoding (
Gcp ortholog encoding (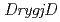 ), YeaZ ortholog (
), YeaZ ortholog ( ), and Gcp ortholog encoding (
), and Gcp ortholog encoding (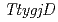 ), YeaZ ortholog (
), YeaZ ortholog (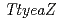 ) in
) in