Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Fujimoto, Hirofumi*; Higuchi, Mariko; Koike, Manabu*; Ode, Hirotaka*; Pinak, M.; Kotulic Bunta, J.*; Nemoto, Toshiyuki*; Sakudo, Takashi*; Honda, Naoko*; Maekawa, Hideaki*; et al.
Journal of Computational Chemistry, 33(3), p.239 - 246, 2012/01
Times Cited Count:36 Percentile:66.42(Chemistry, Multidisciplinary)Lysine acetylation is one of the most common protein post transcriptional modifications. The acetylation effects of lysine residues on Ku protein were examined herein applying several computer simulation techniques. Acetylation of the lysine residues did not reduce the affinity between Ku and its substrate, DNA, in spite of the fact that the substitution of lysine with glutamine (KQ mutant) reduced the affinity of Ku for DNA, or the substitution of lysine with arginine (KR mutant) did not reduce it, as previously reported in experimental studies. These results suggest that the effects of in vivo acetylation may be overestimated when the KQ mutant is employed in mimicry of the acetylated protein.
Higuchi, Mariko; Pinak, M.; Saito, Kimiaki
Hoken Butsuri, 42(2), p.166 - 173, 2007/06
The cluster damaged site on DNA consisting of 7,8-dihydro-8-oxoguanine (8oxoG) and apurinic/apyrimidinic (AP) site is one example of damage that inhibits enzymatic repair. The 2 nanosecond molecular dynamics simulation revealed that the effect of AP site was lager than it of 8oxoG in multiple damaged DNA. The multiple damaged DNA was bent mainly at AP site. Bending direction of multiple damaged DNA including AP site and 8oxoG was different from that at the lesion site of single 8oxoG damaged DNA. These changes may have an influence on the effectiveness of enzymatic repair.
Kotulic Bunta, J.*; Laaksonen, A.*; Pinak, M.; Nemoto, Toshiyuki*
Computational Biology and Chemistry, 30(2), p.112 - 119, 2006/04
Times Cited Count:5 Percentile:30.38(Biology)Due to their lethal consequences, single and double strand breaks are among the most important and dangerous DNA lesions. This work defines and analyzes a DNA with single strand break as a template study for future complex analyses of biologically serious double strand break damage and its enzymatic repair mechanisms. Besides a non-damaged DNA serving as a reference system, system with open valences of the atoms at the strand break ends as well as a system with filled valences was simulated. As for the results, the system with open valences is partly disrupted, and the system with filled valences is stabilized by forming new hydrogen bonds between two strand endings.
Pinak, M.
Modern Methods for Theoretical Physical Chemistry of Biopolymers, p.191 - 210, 2006/00
Results of molecular dynamics (MD) studies of several lesions on DNA and their respective repair enzymes are presented. Main focus is to describe structural and energy changes in DNA molecule with the respect to proper recognition of the lesion by respective repair enzyme. Pyrimidine and purine lesions were studied using MD simulation code Amber and respective force field modified for each lesion. The significant structural changes as breaking of hydrogen bonds network opening and bending the DNA double helix were observed. This collapsing of the double helical structure around the lesion is considered to facilitate docking of the repair enzyme into the DNA. Specific values of electrostatic interaction energy that enables repair enzyme to discriminate lesion from non-damaged site were also detected at several lesion sites.
Fujimoto, Hirofumi*; Pinak, M.; Nemoto, Toshiyuki*; O'Neill, P.*; Kume, Etsuo; Saito, Kimiaki; Maekawa, Hideaki*
Journal of Computational Chemistry, 26(8), p.788 - 798, 2005/06
Times Cited Count:24 Percentile:57.93(Chemistry, Multidisciplinary)Clustered DNA damage sites induced by ionizing radiation have been suggested to have serious consequences to organisms. In this study, approaches based on molecular dynamics (MD) simulation have been applied to examine conformational changes and energetic properties of DNA molecules containing clustered damage sites consisting of 2 lesioned sites, 8-oxoG and AP site. After 1 nanosecond of MD simulation, one of the 6 DNA molecules containing a clustered damage site develop specific characteristic features: sharp bending at the lesioned site and weakening or complete loss of electrostatic interaction energy between 8-oxoG and bases locating on the complementary strand. From these results it is suggested that these changes would make it difficult for the repair enzyme to bind to the lesions within the clustered damage site and thereby result in a reduction of its repair capacity.
Schyman, P.*; Danielsson, J.*; Pinak, M.; Laaksonen, A.*
Journal of Physical Chemistry A, 109(8), p.1713 - 1719, 2005/02
Times Cited Count:40 Percentile:77.11(Chemistry, Physical)We have examined the role of the catalytic lysine (Lys 249) in breaking the glycosidic bond of 8-oxoguanine in the enzyme human 8-oxoguanine DNA glycosylase. It has been assumed that this lysine acts as a nucleophile in a S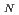 2 type of reaction after being activated through a donation of a proton to a strictly conserved aspartate. We use hybrid density functional theory to characterize both associative and dissociative pathways. We find that the smallest energetical barrier involves a S
2 type of reaction after being activated through a donation of a proton to a strictly conserved aspartate. We use hybrid density functional theory to characterize both associative and dissociative pathways. We find that the smallest energetical barrier involves a S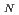 1 type of mechanism where the lysine electrostatically stabilizes the dissociating base and then donates a proton with a very small barrier and then finally attacks the sugar ring to create the covalently bounded protein-DNA intermediate complex. Reported findings give further support to the assumption that a dissociative mechanism may be the preferred mode of action for this type of enzymes.
1 type of mechanism where the lysine electrostatically stabilizes the dissociating base and then donates a proton with a very small barrier and then finally attacks the sugar ring to create the covalently bounded protein-DNA intermediate complex. Reported findings give further support to the assumption that a dissociative mechanism may be the preferred mode of action for this type of enzymes.
Pinak, M.
JAERI 1346, 25 Pages, 2004/12
Molecular dynamics (MD) studies on several radiation damages to DNA - thymine dimer, thymine glycol8-oxoguanine and their recognition by repair enzymes are introduced in order to describe on the stepwise description of molecular process observed at radiation lesion sites. In all cases the significant structural changes in the DNA double helical structure were observed: (a) the breaking of hydrogen bonds network between complementary bases and resulting opening of the double helix (8-oxoguanine); (b) the sharp bending of the DNA helix centered at the lesion site (thymine dimer, thymine glycol); and (c) the flipping-out base on the strand complementary to the lesion (8-oxoguanine). The specific values of electrostatic interaction energy were found at the lesion sites (thymine dimmer: -10kcal/mol, thymine glycol: -26kcal/mol, 8-oxoguanine: -48kcal/mol).
Pinak, M.; O'Neill, P.*; Fujimoto, Hirofumi; Nemoto, Toshiyuki*
AIP Conference Proceedings 708, p.310 - 313, 2004/05
The multiple nanosecond molecular dynamics simulations of DNA mutagenic oxidative lesion - 7,8-dihydro-8-oxoguanine (8-oxoG), complexed with the repair enzyme - human oxoguanine glycosylase 1 (hOGG1) in a physiological aqueous environment, were performed in order to describe the structural and energy changes in DNA and the dynamical process of DNA-enzyme complex formation. In complex the N-terminus of arginine 324 was found located close to the phosphodiester bond of the nucleotide with 8-oxoG enabling chemical reaction(s) between the amino acid and the lesion. The recognition of lesion on DNA, its recognition by repair enzyme and the formation of stable DNA-enzyme complex are necessary conditions for the onset of the successful enzymatic repair process.
Pinak, M.
Hoken Butsuri, 39(1), p.35 - 41, 2004/03
Review of molecular dynamics (MD) studies of several radiation-originated lesions on DNA molecules is presented. Main focus is to describe structural and energy changes in DNA molecule with the respect to proper recognition of the lesion by respective repair enzyme. In most cases the observed changes are related to overall collapsing of the double helical structure around the lesion and are considered to facilitate docking of the repair enzyme into the DNA and formation of DNA-enzyme complex. Stable DNA-enzyme complex is necessary condition for the onset of entire enzymatic repair process. In addition to the structural changes, specific values of electrostatic interaction energy are detected at several lesion sites. The specific electrostatic energy is considered as a factor that enables repair enzyme to discriminate lesion from native, non-damaged site.
Fujimoto, Hirofumi; Pinak, M.; Nemoto, Toshiyuki*; Sakamoto, Kiyotaka*; Yamada, Kazuyuki*; Hoshi, Yoshiyuki*; Kume, Etsuo
Journal of Molecular Structure; THEOCHEM, 681(1-3), p.1 - 8, 2004/01
We developed the novel system, Fujitsu Bio Molecular Visualization System (F-BMVS), that enables to produce real pictures and an animation by arranging them along a time series of a large scale simulation of biomolecules associated with a molecular dynamics (MD) simulation program. This animation system is used to study the results of molecular dynamics code, AMBER, in order to find structural differences on the lesioned DNA comparing with non-damaged DNA. These structural differences would be a factor that guides a repair enzyme to discriminate a lesion from non-damaged DNA region.
Pinak, M.
JAERI-Conf 2003-011, 113 Pages, 2003/09
The workshop "International Workshop on Radiation Risk and its Origin at Molecular and Cellular Level" was held at The Tokai Research Establishment, Japan Atomic Energy Research Institute, on the 6th and 7th of February 2003. The Laboratory of Radiation Risk Analysis of JAERI organized it. This international workshop attracted scientists from several different scientific areas, including radiation physics, radiation biology, molecular biology, crystallography of biomolecules, modeling and bio-informatics. Several foreign and domestic keynote speakers addresses the very fundamental areas of radiation risk and tried to establish a link between the fundamental studies at the molecular and cellular level and radiation damages at the organism. The symposium consisted of 13 oral lectures, 10 poster presentations and panel discussion. The 108 participants attended the workshop. This publication comprises of proceedings of oral and poster presentations where available. For the rest of contributions the abstracts or/and selections of presentation materials are shown instead.
Pinak, M.
Computational Biology and Chemistry, 27(3), p.431 - 441, 2003/07
Times Cited Count:9 Percentile:44.66(Biology)One nanosecond molecular dynamics (MD) simulation was performed for two DNA segments each composed of 30 base pairs. The analysis of results was focused on the electrostatic energy that is supposed to be significant factor causing the disruption of DNA base stacking in DNA duplex and may also to serve as a signal toward the repair enzyme informing on the presence of the lesion. This electrostatic potential may signal presence of the lesion to the repair enzyme.
Pinak, M.
Journal of Computational Chemistry, 24(7), p.898 - 907, 2003/04
Times Cited Count:8 Percentile:35.73(Chemistry, Multidisciplinary)The molecular dynamics (MD) simulation of DNA mutagenic oxidative lesion 8-oxoG, complexed with the repair enzyme - hOGG1 was performed in order to describe the dynamical process of DNA-enzyme complex formation. After 500 picoseconds of MD the lesioned DNA and enzyme formed a complex that lasted stable until the simulation was terminated at 1 ns. The N-terminus of arginine 313 was located close to the phosphodiester bond of nucleotide with 8-oxoG enabling chemical reactions between amino acid and lesion. Phosphodiester bond at C5' of 8-oxoG was displaced to the position close to the N-terminus of arginine 313. The water mediated hydrogen bonds network was formed in each contact area between DNA and enzyme further enhancing the stability of complex.
Pinak, M.
Central European Journal of Physics, 1(1), p.179 - 190, 2003/01
The molecular dynamics (MD) studies of several radiation originated lesions on the DNA molecules are presented with the respect to the proper recognition of the lesion by the respective repair enzyme. Several pyrimidine and purine lesions were subjected to the MD simulations for several hundreds picoseconds using MD simulation code AMBER 5.0 and respective force field modified for the lesion. In all cases the significant structural changes in the DNA double helical structure were observed. These changes were related to the overall collapsing double helical structure around the lesion and are supposed to facilitate the docking of the repair enzyme into the DNA in formation of DNA-enzyme complex. In addition to the structural changes, the specific values of electrostatic interaction energy were found at several lesion sites.
Pinak, M.; Laaksonen, A.
Molecular Mechanisms for Radiation-Induced Cellular Response and Cancer Development, p.266 - 273, 2003/00
The molecular dynamics (MD) simulations of DNA mutagenic oxidative lesion - 7,8-dihydro-8-oxoguanine (8-oxoG), single and complexed with the repair enzyme - human oxoguanine glycosylase 1 (hOGG1), were performed for 1 nanosecond (ns) in order to determine structural changes at the DNA molecule and to describe a dynamical process of DNA-enzyme complex formation. The broken hydrogen bonds resulting in locally collapsed B-DNA structure were observed at the lesion site. In addition the adenine on the complementary strand (separated from 8-oxoG by 1 base pair) was flipped-out of the DNA double helix. In the case of simulation of DNA and repair enzyme hOGG1, the DNA-enzyme complex was formed after 500 picoseconds of MD that lasted stable until the simulation was terminated at 1 ns.
Pinak, M.; Nemoto, Toshiyuki*
Proceedings of International Conference on Supercomputing in Nuclear Applications (SNA 2003) (CD-ROM), 15 Pages, 2003/00
The molecular dynamics (MD) studies of oxidative lesions on DNA molecules are presented with respect to the proper recognition of lesions by their respective repair enzymes. The recognition of lesion and the formation of stable DNA-enzyme complex are necessary conditions for the onset of the enzymatic repair process. Two DNA lesions, thymine glycol and 8-oxoguanine were subjected to the MD simulations for several hundreds picoseconds using MD simulation codes AMBER 5.0 and AMBER 7.0. Amber was changed and partly vectorized to be able to handle this large systems. Simulations were performed on FUJITSU VPP5000/64 vector/parallel type, the HITACHI SR8000 and FUJITSU PRIMEPOWER parallel types supercomputers.
Pinak, M.
JAERI-Research 2002-016, 31 Pages, 2002/09
The molecular dynamics (MD) simulations of DNA mutagenic oxidative lesion 8-oxoG, single and complexed with the repair enzyme hOGG1, were performed. In the case of simulation of single DNA molecule the broken hydrogen bonds resulting in locally collapsed B-DNA structure were observed at the lesion site. In addition the adenine on the complementary strand was flipped-out of the DNA double helix. In the case of simulation of DNA and repair enzyme hOGG1, the DNA-enzyme complex was formed after 500 picoseconds of MD that lasted stable until the simulation was terminated at 1 ns. The N-terminus of arginine 313 was located close to the phosphodiester bond of nucleotide with 8-oxoG enabling chemical reactions between amino acid and lesion. Phosphodiester bond at C5'of 8-oxoG was at the position close to the N-terminus of arginine 313. The water mediated hydrogen bonds network was formed in each contact area between DNA and enzyme further enhancing the stability of complex.
Pinak, M.
Journal of Molecular Structure; THEOCHEM, 583(1-3), p.189 - 197, 2002/04
The local structural and energetic impact of a mutagenic oxidative lesion 7,8-dihydro-8-oxoguanine (8-oxoG) on a DNA molecule was studied by the method of a molecular dynamics (MD) simulation. The molecule of 8-oxoG was inserted into central part of B-DNA 15-mer d(GCGTCCA'8-oxoG'GTCTACC) replacing the native guanine. The 2-nanosecond MD simulations were performed with the AMBER 5.0 program code at the constant temperature of 310 K (
replacing the native guanine. The 2-nanosecond MD simulations were performed with the AMBER 5.0 program code at the constant temperature of 310 K ( 36.5ºC, temperature of human body) for the 8-oxoG lesioned and native DNA molecules. The broken hydrogen bonds resulting in locally collapsed B-DNA structure were observed at the lesion site. The adenine 21 on the complementary strand (separated from 8-oxoG by 1 base pair) is flipped-out of the DNA double helix. Its extrahelical position forms a hole that may favor docking of repair enzyme into DNA during repair process. A strong electrostatic repulsion between nucleotide with the 8-oxoG and neighboring nucleotides contributes to the observed instability of DNA at the lesion.
36.5ºC, temperature of human body) for the 8-oxoG lesioned and native DNA molecules. The broken hydrogen bonds resulting in locally collapsed B-DNA structure were observed at the lesion site. The adenine 21 on the complementary strand (separated from 8-oxoG by 1 base pair) is flipped-out of the DNA double helix. Its extrahelical position forms a hole that may favor docking of repair enzyme into DNA during repair process. A strong electrostatic repulsion between nucleotide with the 8-oxoG and neighboring nucleotides contributes to the observed instability of DNA at the lesion.
Pinak, M.
JAERI-Conf 2002-005, 201 Pages, 2002/03
The Book of Proceedings comprises of proceedings (or abstracts), program and list of participants from the Workshop "Recognition of DNA damage as onset of successful repair: Computational and experimental approaches" that was organized by Radiation Risk Analysis Laboratory and held on December 18-19, 2001 in JAERI, Tokai. The workshop proceedings consist of researches from the following areas: Radiation interaction with DNA and its environment; Detection of DNA mutations; Structural biology & computing; DNA damage & enzymatic repair; and DNA imaging.
Pinak, M.
Journal of Computational Chemistry, 22(15), p.1723 - 1731, 2001/11
Times Cited Count:3 Percentile:21.81(Chemistry, Multidisciplinary)no abstracts in English