Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Papadopoulos, A.*; Kyriakou, I.*; Matsuya, Yusuke; Cort s-Giraldo, M. A.*; Galocha-Oliva, M.*; Plante, I.*; Steward, R. D.*; Tran, N. H.*; Li, W.*; Daglis, I. A.*; et al.
s-Giraldo, M. A.*; Galocha-Oliva, M.*; Plante, I.*; Steward, R. D.*; Tran, N. H.*; Li, W.*; Daglis, I. A.*; et al.
Radiation and Environmental Biophysics, 64(1), p.117 - 135, 2025/03
Radiation quality for determining biological effects is commonly linked to the microdosimetric quantity, especially dose-mean lineal energy y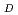 . Calculations of y
. Calculations of y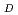 can be performed by sophisticated Monte Carlo track structure (MCTS) codes. The y
can be performed by sophisticated Monte Carlo track structure (MCTS) codes. The y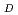 estimate depends on the type of the MCTS code and analysis model. This study focused on proton beams with 1 MeV-1 GeV, which are important in radiation protection, space applications, radiation therapy, etc., and compared the estimates of the y
estimate depends on the type of the MCTS code and analysis model. This study focused on proton beams with 1 MeV-1 GeV, which are important in radiation protection, space applications, radiation therapy, etc., and compared the estimates of the y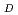 in liquid water by various MCTS codes (PHITS, RITRACK, and Geant4-DNA) and analysis models (refined Xapsos model). The comparison results showed that good agreement with the refined Xapsos model and various MCTS codes can be attained at less than 10-20% level, and Q values by the analytic model are also in better agreement with MCTS simulation data. These findings conclude that the refined analytic model might be used as an alternative to time- and CPU-intensive MCTS simulations and advance practical calculations of radiation qualities and risk assessment.
in liquid water by various MCTS codes (PHITS, RITRACK, and Geant4-DNA) and analysis models (refined Xapsos model). The comparison results showed that good agreement with the refined Xapsos model and various MCTS codes can be attained at less than 10-20% level, and Q values by the analytic model are also in better agreement with MCTS simulation data. These findings conclude that the refined analytic model might be used as an alternative to time- and CPU-intensive MCTS simulations and advance practical calculations of radiation qualities and risk assessment.
Polster, L.*; Schuemann, J.*; Rinaldi, I.*; Burigo, L.*; McNamara, A. L.*; Steward, R. D.*; Attili, A.*; Carlson, D. J.*; Sato, Tatsuhiko; Ramos M ndez, J.*; et al.
ndez, J.*; et al.
Physics in Medicine & Biology, 60(13), p.5053 - 5070, 2015/07
Times Cited Count:55 Percentile:90.25(Engineering, Biomedical)The aim of this work is to extend a widely used proton Monte Carlo tool, TOPAS, towards the modeling of relative biological effect (RBE) distributions in experimental arrangements as well as patients. Then, eight biophysical models was incorporated into TOPAS. As far as physics parameters are concerned, four of these models are based on the proton linear energy transfer (LET), while the others are based on DNA Double Strand Break (DSB) induction and the frequency-mean specific energy, lineal energy, or delta electron generated track structure. The model on the basis of lineal energy adapted a microdosimetric function developed by JAEA. This work is an important step in bringing biologically optimized treatment planning for proton therapy closer to the clinical practice.