Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Watanabe, Tomoaki; Suyama, Kenya; Tada, Kenichi; Ferrer, R. M.*; Hykes, J.*; Wemple, C. A.*
Nuclear Science and Engineering, 198(11), p.2230 - 2239, 2024/11
Times Cited Count:1 Percentile:51.66(Nuclear Science & Technology)A new nuclear data library for the advanced lattice physics code CASMO5 has been prepared based on JENDL-5. In JENDL-5, many essential nuclides for conventional LWR analysis have also been modified based on state-of-the-art evaluations. The new JENDL-5-based CASMO5 library was prepared by replacing as much of the nuclear data of the current CASMO5 ENDF/B-VII.1-based library as possible with JENDL-5. This study verified and validated the new library. Verifications were performed based on the OECD/NEA burnup credit criticality safety benchmark phase III-C, and the calculated k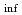 and fuel compositions of the BWR fuel assembly were compared with reported benchmark results. Comparison with the MCNP6.2 result was also performed using the same benchmark model. In addition, the TCA critical experiment and Takahama-3 post-irradiation experiment were used for validation. The results indicate that the new library performs well and is comparable to the ENDF/B-VII.1-based library in predictions of reactivity and fuel compositions for LWR systems.
and fuel compositions of the BWR fuel assembly were compared with reported benchmark results. Comparison with the MCNP6.2 result was also performed using the same benchmark model. In addition, the TCA critical experiment and Takahama-3 post-irradiation experiment were used for validation. The results indicate that the new library performs well and is comparable to the ENDF/B-VII.1-based library in predictions of reactivity and fuel compositions for LWR systems.
Yamasaki, Chisato*; Murakami, Katsuhiko*; Fujii, Yasuyuki*; Sato, Yoshiharu*; Harada, Erimi*; Takeda, Junichi*; Taniya, Takayuki*; Sakate, Ryuichi*; Kikugawa, Shingo*; Shimada, Makoto*; et al.
Nucleic Acids Research, 36(Database), p.D793 - D799, 2008/01
Times Cited Count:52 Percentile:70.47(Biochemistry & Molecular Biology)Here we report the new features and improvements in our latest release of the H-Invitational Database, a comprehensive annotation resource for human genes and transcripts. H-InvDB, originally developed as an integrated database of the human transcriptome based on extensive annotation of large sets of fulllength cDNA (FLcDNA) clones, now provides annotation for 120 558 human mRNAs extracted from the International Nucleotide Sequence Databases (INSD), in addition to 54 978 human FLcDNAs, in the latest release H-InvDB. We mapped those human transcripts onto the human genome sequences (NCBI build 36.1) and determined 34 699 human gene clusters, which could define 34 057 protein-coding and 642 non-protein-coding loci; 858 transcribed loci overlapped with predicted pseudogenes.
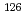 Cs(
Cs(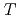
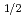 = 1.63 m) by ABMR
= 1.63 m) by ABMRPinard, J.*; Duong, H. T.*; Marescaux, D.*; Stroke, H. H.*; Redi, O.*; Gustafsson, M.*; Nilsson, T.*; Matsuki, S.*; Kishimoto, Yasuaki*; Kominato, K.*; et al.
Nuclear Physics A, 753(1-2), p.3 - 12, 2005/05
Times Cited Count:4 Percentile:35.32(Physics, Nuclear)The hfs separation 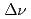 of
of 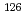 Cs(
Cs(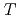
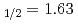 m) in the 6s
m) in the 6s 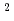 S
S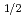 ground state was obtained in a precision measurement near zero magnetic field by means of atomic beam magnetic resonance with laser optical pumping on-line with the CERN-PSB-ISOLDE mass separator. The result,
ground state was obtained in a precision measurement near zero magnetic field by means of atomic beam magnetic resonance with laser optical pumping on-line with the CERN-PSB-ISOLDE mass separator. The result, 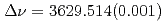 MHz, corrects significantly a previous published value from a high-field experiment. With our result, the precision of the nuclear magnetic moment,
MHz, corrects significantly a previous published value from a high-field experiment. With our result, the precision of the nuclear magnetic moment, 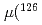 Cs)
Cs)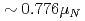 , is now limited by the influence of extended nuclear structure on the hfs (the Bohr-Weisskopf effect).
, is now limited by the influence of extended nuclear structure on the hfs (the Bohr-Weisskopf effect).
 -ray irradiation
-ray irradiationG.M.Qin*; ; Hatada, Motoyoshi
JAERI-M 91-106, 114 Pages, 1991/07
no abstracts in English
Glugla, M.*; Beloglazov, S.*; Carlson, B.*; Cho, S.*; Cristescu, I.*; Cristecu, I.*; Chung, H.*; Girard, J.-P.*; Hayashi, Takumi; Mardoch, D.*; et al.
no journal, ,