Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Al-Shayeb, B.*; Sachdeva, R.*; Chen, L.-X.*; Ward, F.*; Munk, P.*; Devoto, A.*; Castelle, C. J.*; Olm, M. R.*; Bouma-Gregson, K.*; Amano, Yuki; et al.
Nature, 578(7795), p.425 - 431, 2020/02
Times Cited Count:295 Percentile:99.46(Multidisciplinary Sciences)Matheus Carnevali, P. B.*; Schulz, F.*; Castelle, C. J.*; Kantor, R. S.*; Shih, P.*; Sharon, I.*; Santini, J.*; Olm, M. R.*; Amano, Yuki; Thomas, B. C.*; et al.
Nature Communications (Internet), 10, p.463_1 - 463_15, 2019/01
Times Cited Count:48 Percentile:88.33(Multidisciplinary Sciences)Bandodkar, A. J.*; Gutruf, P.*; Choi, J.*; Lee, K.-H.*; Sekine, Yurina; Reeder, J. T.*; Jeang, W. J.*; Aranyosi, A. J.*; Lee, S. P.*; Model, J. B.*; et al.
Science Advances (Internet), 5(1), p.eaav3294_1 - eaav3294_15, 2019/01
Times Cited Count:566 Percentile:99.88(Multidisciplinary Sciences)Interest in advanced wearable technologies increasingly extends beyond systems for biophysical measurements to those that enable continuous, non-invasive monitoring of biochemical markers in biofluids. Here, we introduce battery-free, wireless microelectronic platforms that perform sensing via schemes inspired by the operation of biofuel cells. Combining these systems in a magnetically releasable manner with chrono-sampling microfluidic networks that incorporate assays based on colorimetric sensing yields thin, flexible, lightweight, skin-interfaced technologies with broad functionality in sweat analysis. A demonstration device allows simultaneous monitoring of sweat rate/loss, along with quantitative measurements of pH and of lactate, glucose and chloride concentrations using biofuel cell and colorimetric approaches.
Ino, Kohei*; Hernsdorf, A. W.*; Konno, Yuta*; Kozuka, Mariko*; Yanagawa, Katsunori*; Kato, Shingo*; Sunamura, Michinari*; Hirota, Akinari*; Togo, Yoko*; Ito, Kazumasa*; et al.
ISME Journal, 12(1), p.31 - 47, 2018/01
Times Cited Count:54 Percentile:89.82(Ecology)In this study, we found the dominance ofanaerobic methane-oxidizing archaea in groundwater enriched in sulfate and methane from a 300-m deep underground borehole in granitic rock.
 -spectroscopy at an intense cold neutron beam facility
-spectroscopy at an intense cold neutron beam facilityJentschel, M.*; Blanc, A.*; de France, G.*; K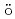 ster, U.*; Leoni, S.*; Mutti, P.*; Simpson, G.*; Soldner, T.*; Ur, C.*; Urban, W.*; et al.
ster, U.*; Leoni, S.*; Mutti, P.*; Simpson, G.*; Soldner, T.*; Ur, C.*; Urban, W.*; et al.
Journal of Instrumentation (Internet), 12(11), p.P11003_1 - P11003_33, 2017/11
Times Cited Count:43 Percentile:84.98(Instruments & Instrumentation) and metal transformations associated with novel bacteria and archaea in deep terrestrial subsurface sediments
and metal transformations associated with novel bacteria and archaea in deep terrestrial subsurface sedimentsHernsdorf, A. W.*; Amano, Yuki; Miyakawa, Kazuya; Ise, Kotaro; Suzuki, Yohei*; Anantharaman, K.*; Probst, A. J.*; Burstein, David*; Thomas, B. C.*; Banfield, J. F.*
ISME Journal, 11, p.1915 - 1929, 2017/03
Times Cited Count:107 Percentile:96.15(Ecology)To evaluate the potential for interactions between microbial communities and disposal systems, we explored the structure and metabolic function of a sediment-hosted subsurface ecosystem associated with Horonobe Underground Research Center, Hokkaido, Japan. Overall, the ecosystem is enriched in organisms from diverse lineages and many are from phyla that lack isolated representatives. The majority of organisms can metabolize H , often via oxidative [NiFe] hydrogenases or electron-bifurcating [FeFe] hydrogenases that enable ferredoxin-based pathways, including the ion motive Rnf complex. Many organisms implicated in H
, often via oxidative [NiFe] hydrogenases or electron-bifurcating [FeFe] hydrogenases that enable ferredoxin-based pathways, including the ion motive Rnf complex. Many organisms implicated in H metabolism are also predicted to catalyze carbon, nitrogen, iron, and sulfur transformations. Notably, iron-based metabolism was predicted in a bacterial lineage where this function has not been predicted previously and in an ANME-2d archaeaon that is implicated in methane oxidation. We infer an ecological model that links microorganisms to sediment-derived resources and predict potential impacts of microbial activity on H
metabolism are also predicted to catalyze carbon, nitrogen, iron, and sulfur transformations. Notably, iron-based metabolism was predicted in a bacterial lineage where this function has not been predicted previously and in an ANME-2d archaeaon that is implicated in methane oxidation. We infer an ecological model that links microorganisms to sediment-derived resources and predict potential impacts of microbial activity on H accumulation and radionuclide migration.
accumulation and radionuclide migration.
Hug, L. A.*; Baker, B. J.*; Anantharaman, K.*; Brown, C. T.*; Probst, A. J.*; Castelle, C. J.*; Butterfield, C. N.*; Hernsdorf, A. W.*; Amano, Yuki; Ise, Kotaro; et al.
Nature Microbiology (Internet), 1(5), p.16048_1 - 16048_6, 2016/05
Times Cited Count:1378 Percentile:99.96(Microbiology)The tree of life is one of the most important organizing principles in biology. Gene surveys suggest the existence of an enormous number of branches, but even an approximation of the full scale of The Tree has remained elusive. Here, we use newly available information from genomes of uncultivated organisms, along with other published sequences, to present a new version of the Tree of life, with Bacteria, Archaea and Eukaryotes included. The depiction is both a global overview and a snapshot of the diversity within each major lineage. The results imply the predominance of bacterial diversification and underline the importance of organisms lacking isolated representatives, with substantial evolution concentrated in a major radiation of such organisms.
Bulanov, S. S.*; Chen, M.*; Schroeder, C. B.*; Esarey, E.*; Leemans, W. P.*; Bulanov, S. V.; Esirkepov, T. Z.; Kando, Masaki; Koga, J. K.; Zhidkov, A. G.*; et al.
AIP Conference Proceedings 1507, p.825 - 830, 2012/12
Times Cited Count:9 Percentile:92.14(Physics, Applied)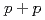 collisions at
collisions at 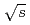 = 200 GeV
= 200 GeVAdare, A.*; Afanasiev, S.*; Aidala, C.*; Ajitanand, N. N.*; Akiba, Y.*; Al-Bataineh, H.*; Alexander, J.*; Aoki, K.*; Aphecetche, L.*; Armendariz, R.*; et al.
Physical Review D, 84(1), p.012006_1 - 012006_18, 2011/07
Times Cited Count:33 Percentile:75.18(Astronomy & Astrophysics)We report on the event structure and double helicity asymmetry (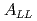 ) of jet production in longitudinally polarized
) of jet production in longitudinally polarized 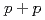 collisions at
collisions at 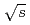 = 200 GeV. Photons and charged particles were measured by the PHENIX experiment. Event structure was compared with the results from PYTHIA event generator. The production rate of reconstructed jets is satisfactorily reproduced with the next-to-leading-order perturbative QCD calculation. We measured
= 200 GeV. Photons and charged particles were measured by the PHENIX experiment. Event structure was compared with the results from PYTHIA event generator. The production rate of reconstructed jets is satisfactorily reproduced with the next-to-leading-order perturbative QCD calculation. We measured 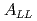 = -0.0014
= -0.0014 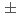 0.0037 at the lowest
0.0037 at the lowest 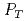 bin and -0.0181
bin and -0.0181 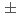 0.0282 at the highest
0.0282 at the highest 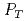 bin. The measured
bin. The measured 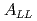 is compared with the predictions that assume various
is compared with the predictions that assume various 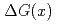 distributions.
distributions.
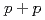 collisions at
collisions at 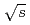 = 200 and 62.4 GeV
= 200 and 62.4 GeVAdare, A.*; Afanasiev, S.*; Aidala, C.*; Ajitanand, N. N.*; Akiba, Yasuyuki*; Al-Bataineh, H.*; Alexander, J.*; Aoki, Kazuya*; Aphecetche, L.*; Armendariz, R.*; et al.
Physical Review C, 83(6), p.064903_1 - 064903_29, 2011/06
Times Cited Count:193 Percentile:99.41(Physics, Nuclear)Transverse momentum distributions and yields for 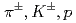 , and
, and 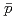 in
in 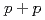 collisions at
collisions at 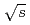 = 200 and 62.4 GeV at midrapidity are measured by the PHENIX experiment at the RHIC. We present the inverse slope parameter, mean transverse momentum, and yield per unit rapidity at each energy, and compare them to other measurements at different
= 200 and 62.4 GeV at midrapidity are measured by the PHENIX experiment at the RHIC. We present the inverse slope parameter, mean transverse momentum, and yield per unit rapidity at each energy, and compare them to other measurements at different 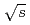 collisions. We also present the scaling properties such as
collisions. We also present the scaling properties such as 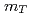 and
and 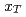 scaling and discuss the mechanism of the particle production in
scaling and discuss the mechanism of the particle production in 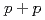 collisions. The measured spectra are compared to next-to-leading order perturbative QCD calculations.
collisions. The measured spectra are compared to next-to-leading order perturbative QCD calculations.
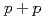 and Au+Au collisions at
and Au+Au collisions at 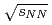 = 200 GeV
= 200 GeVAdare, A.*; Afanasiev, S.*; Aidala, C.*; Ajitanand, N. N.*; Akiba, Yasuyuki*; Al-Bataineh, H.*; Alexander, J.*; Aoki, Kazuya*; Aphecetche, L.*; Aramaki, Y.*; et al.
Physical Review C, 83(4), p.044912_1 - 044912_16, 2011/04
Times Cited Count:10 Percentile:54.53(Physics, Nuclear)Measurements of electrons from the decay of open-heavy-flavor mesons have shown that the yields are suppressed in Au+Au collisions compared to expectations from binary-scaled 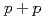 collisions. Here we extend these studies to two particle correlations where one particle is an electron from the decay of a heavy flavor meson and the other is a charged hadron from either the decay of the heavy meson or from jet fragmentation. These measurements provide more detailed information about the interaction between heavy quarks and the quark-gluon matter. We find the away-side-jet shape and yield to be modified in Au+Au collisions compared to
collisions. Here we extend these studies to two particle correlations where one particle is an electron from the decay of a heavy flavor meson and the other is a charged hadron from either the decay of the heavy meson or from jet fragmentation. These measurements provide more detailed information about the interaction between heavy quarks and the quark-gluon matter. We find the away-side-jet shape and yield to be modified in Au+Au collisions compared to 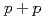 collisions.
collisions.
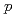 +
+ 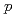 collisions at
collisions at 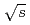 = 200 GeV and scaling properties of hadron production
= 200 GeV and scaling properties of hadron productionAdare, A.*; Afanasiev, S.*; Aidala, C.*; Ajitanand, N. N.*; Akiba, Y.*; Al-Bataineh, H.*; Alexander, J.*; Aoki, K.*; Aphecetche, L.*; Armendariz, R.*; et al.
Physical Review D, 83(5), p.052004_1 - 052004_26, 2011/03
Times Cited Count:181 Percentile:98.32(Astronomy & Astrophysics)The PHENIX experiment at RHIC has measured the invariant differential cross section for production of 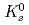 ,
, 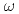 ,
, 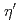 and
and 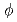 mesons in
mesons in 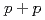 collisions at
collisions at 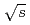 = 200 GeV. The spectral shapes of all hadron transverse momentum distributions are well described by a Tsallis distribution functional form with only two parameters,
= 200 GeV. The spectral shapes of all hadron transverse momentum distributions are well described by a Tsallis distribution functional form with only two parameters, 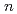 and
and 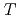 , determining the high
, determining the high 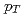 and characterizing the low
and characterizing the low 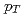 regions for the spectra, respectively. The integrated invariant cross sections calculated from the fitted distributions are found to be consistent with existing measurements and with statistical model predictions.
regions for the spectra, respectively. The integrated invariant cross sections calculated from the fitted distributions are found to be consistent with existing measurements and with statistical model predictions.
Walsh, M.*; Andrew, P.*; Barnsley, R.*; Bertalot, L.*; Boivin, R.*; Bora, D.*; Bouhamou, R.*; Ciattaglia, S.*; Costley, A. E.*; Counsell, G.*; et al.
Proceedings of 23rd IAEA Fusion Energy Conference (FEC 2010) (CD-ROM), 8 Pages, 2011/03
Reichle, R.*; Andrew, P.*; Counsell, G.*; Drevon, J.-M.*; Encheva, A.*; Janeschitz, G.*; Johnson, D. W.*; Kusama, Yoshinori; Levesy, B.*; Martin, A.*; et al.
Review of Scientific Instruments, 81(10), p.10E135_1 - 10E135_5, 2010/10
Times Cited Count:35 Percentile:77.43(Instruments & Instrumentation)Yamasaki, Chisato*; Murakami, Katsuhiko*; Fujii, Yasuyuki*; Sato, Yoshiharu*; Harada, Erimi*; Takeda, Junichi*; Taniya, Takayuki*; Sakate, Ryuichi*; Kikugawa, Shingo*; Shimada, Makoto*; et al.
Nucleic Acids Research, 36(Database), p.D793 - D799, 2008/01
Times Cited Count:52 Percentile:70.47(Biochemistry & Molecular Biology)Here we report the new features and improvements in our latest release of the H-Invitational Database, a comprehensive annotation resource for human genes and transcripts. H-InvDB, originally developed as an integrated database of the human transcriptome based on extensive annotation of large sets of fulllength cDNA (FLcDNA) clones, now provides annotation for 120 558 human mRNAs extracted from the International Nucleotide Sequence Databases (INSD), in addition to 54 978 human FLcDNAs, in the latest release H-InvDB. We mapped those human transcripts onto the human genome sequences (NCBI build 36.1) and determined 34 699 human gene clusters, which could define 34 057 protein-coding and 642 non-protein-coding loci; 858 transcribed loci overlapped with predicted pseudogenes.
Amano, Yuki; Diamond, S.*; Lavy, A.*; Anantharaman, K.*; Miyakawa, Kazuya; Iwatsuki, Teruki; Beppu, Hikari*; Suzuki, Yohei*; Thomas, B. C.*; Banfield, J. F.*
no journal, ,
Amano, Yuki; Sachdeva, R.*; Gittins, D.*; Anantharaman, K.*; Lei, S.*; Beppu, Hikari*; Mochizuki, Akihito; Thomas, B. C.*; Suzuki, Yohei*; Banfield, J. F.*
no journal, ,
Amano, Yuki; Beppu, Hikari*; Endo, Takashi*; Nemoto, Kazuaki*; Sato, Tomofumi*; Thomas, B. C.*; Banfield, J. F.*
no journal, ,
Amano, Yuki; Beppu, Hikari*; Sato, Tomofumi*; Mochizuki, Akihito; Thomas, B. C.*; Banfield, J. F.*
no journal, ,