Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Inoue, Rintaro*; Oda, Takashi; Nakagawa, Hiroshi; Tominaga, Taiki*; Ikegami, Takahisa*; Konuma, Tsuyoshi*; Iwase, Hiroki*; Kawakita, Yukinobu; Sato, Mamoru*; Sugiyama, Masaaki*
Biophysical Journal, 124(3), p.540 - 548, 2025/02
Times Cited Count:0 Percentile:0.00(Biophysics)Kim, J.*; Thompson, B. R.*; Tominaga, Taiki*; Osawa, Takahito; Egami, Takeshi*; F rster, S.*; Ohl, M.*; Senses, E.*; Faraone, A.*; Wagner, N. J.*
rster, S.*; Ohl, M.*; Senses, E.*; Faraone, A.*; Wagner, N. J.*
ACS Macro Letters (Internet), 13(6), p.720 - 725, 2024/06
Times Cited Count:1 Percentile:28.80(Polymer Science)The Rouse dynamics of polymer chains in model nanocomposite PolyEthylene Oxide (PEO)/Silica NanoParticles (NPs) was investigated using QuasiElastic Neutron Scattering (QENS). The apparent Rouse rate of the polymer chains decreases as the particle loading increases. However, there is no evidence of an immobile segment population on the probed time scale of tens of ps. The slowing down of the dynamics is interpreted in terms of modified Rouse models for the chains in the NP inter-phase region. Thus, two chain populations, one bulk like and the other characterized by a suppression of Rouse modes, are identified. The spatial extent of the interphase region is estimated to be about twice the adsorbed layer thickness, or about 2 nm. These findings provide a detailed description of the suppression of the chain dynamics on the surface of NPs. These results are relevant insights on surface effects and confinement and provide a foundation for the understanding of the rheological properties of Polymer NanoComposites (PNCs) with well-dispersed NPs.
Hirota, Yuki*; Tominaga, Taiki*; Kawabata, Takashi*; Kawakita, Yukinobu; Matsuo, Yasumitsu*
Bioengineering (Internet), 10(5), p.622_1 - 622_17, 2023/05
Times Cited Count:3 Percentile:38.63(Biotechnology & Applied Microbiology)Hirota, Yuki*; Tominaga, Taiki*; Kawabata, Takashi*; Kawakita, Yukinobu; Matsuo, Yasumitsu*
Bioengineering (Internet), 9(10), p.599_1 - 599_17, 2022/10
Times Cited Count:3 Percentile:22.64(Biotechnology & Applied Microbiology)Tominaga, Taiki*; Nakagawa, Hiroshi; Sahara, Masae*; Oda, Takashi*; Inoue, Rintaro*; Sugiyama, Masaaki*
Life (Internet), 12(5), p.675_1 - 675_9, 2022/05
Times Cited Count:2 Percentile:20.29(Biology)The background scattering of sample cells suitable for aqueous protein solution samples, conducted with a neutron backscattering spectrometer, was evaluated. It was found that the scattering intensity of an aluminum sample cell coated with boehmite using D O was lower than that of a sample cell coated with regular water (H
O was lower than that of a sample cell coated with regular water (H O). In addition, meticulous attention to cells with small individual weight differences and the positional reproducibility of the sample cell relative to the spectrometer neutron beam position enabled the accurate subtraction of the scattering profiles of the D
O). In addition, meticulous attention to cells with small individual weight differences and the positional reproducibility of the sample cell relative to the spectrometer neutron beam position enabled the accurate subtraction of the scattering profiles of the D O buffer and the sample container. Consequently, high quality information on protein dynamics could be extracted from dilute protein solutions.
O buffer and the sample container. Consequently, high quality information on protein dynamics could be extracted from dilute protein solutions.
Furuike, Yoshihiko*; Ouyang, D.*; Tominaga, Taiki*; Matsuo, Tatsuhito*; Mukaiyama, Atsushi*; Kawakita, Yukinobu; Fujiwara, Satoru*; Akiyama, Shuji*
Communications Physics (Internet), 5(1), p.75_1 - 75_12, 2022/04
Times Cited Count:7 Percentile:63.47(Physics, Multidisciplinary)Tominaga, Taiki*; Sahara, Masae*; Kawakita, Yukinobu; Nakagawa, Hiroshi; Yamada, Takeshi*
Journal of Applied Crystallography, 54(6), p.1631 - 1640, 2021/12
Times Cited Count:6 Percentile:57.51(Chemistry, Multidisciplinary)Nogami, Satoshi*; Kadota, Kazunori*; Uchiyama, Hiromasa*; Arima-Osonoi, Hiroshi*; Iwase, Hiroki*; Tominaga, Taiki*; Yamada, Takeshi*; Takata, Shinichi; Shibayama, Mitsuhiro*; Tozuka, Yuichi*
International Journal of Biological Macromolecules, 190, p.989 - 998, 2021/11
Times Cited Count:9 Percentile:45.87(Biochemistry & Molecular Biology)Nakagawa, Hiroshi; Saio, Tomohide*; Nagao, Michihiro*; Inoue, Rintaro*; Sugiyama, Masaaki*; Ajito, Satoshi; Tominaga, Taiki*; Kawakita, Yukinobu
Biophysical Journal, 120(16), p.3341 - 3354, 2021/08
Times Cited Count:7 Percentile:43.91(Biophysics)A multi-domain protein can have various conformations in solution. Interactions with other molecules result in the stabilization of one of the conformations and change in the domain dynamics. SAXS, a well-established experimental technique, can be employed to elucidate the conformation of a multi-domain protein in solution. NSE spectroscopy is a promising technique for recording the domain dynamics in nanosecond and nanometer scale. Despite the great efforts, there are still under development. Thus, we quantitatively removed the contribution of diffusion dynamics and hydrodynamic interactions from the NSE data via incoherent scattering, revealing the differences in the domain dynamics of the three functional states of a multi-domain protein, MurD. The differences among the three states can be explained by two domain modes.
Matsuura, Masato*; Yamada, Takeshi*; Tominaga, Taiki*; Kobayashi, Makoto*; Nakagawa, Hiroshi; Kawakita, Yukinobu
JPS Conference Proceedings (Internet), 33, p.011068_1 - 011068_6, 2021/03
The position dependence of the scattered intensity in the time-of-flight backscattering spectrometer DNA was investigated. A periodic structure for both vertical (pixel) and horizontal (PSD) directions was observed. The solar slit and over-bending of an analyzer crystal is discussed as a possible origin of the modulation in the intensity. We have developed software program for the systematic correction of the position-dependent intensity and offset energy for the elastic peak. This corrects the deviation from the true scattering intensity and improve the quality of the data, which includes the energy resolution.
Tominaga, Taiki*; Kobayashi, Makoto*; Yamada, Takeshi*; Matsuura, Masato*; Kawakita, Yukinobu; Kasai, Satoshi*
JPS Conference Proceedings (Internet), 33, p.011095_1 - 011095_5, 2021/03
A vertical movement type of sample changer for the neutron spectrometer BL02, J-PARC MLF was developed for our top-loading type cryostat. The sample changer, termed as "PEACE", can control reproducibility of the irradiated position using guides made of polyether ether ketone. The variation between the background scattering profiles of three sample positions was found to be less than plus minus 1.6%. This result is reasonable, considering the deviation of sample position of less than plus minus 0.3 mm from the vertical axis.
Tominaga, Taiki*; Sahara, Masae*; Kawakita, Yukinobu; Nakagawa, Hiroshi; Shimamoto, Naonobu*
JPS Conference Proceedings (Internet), 33, p.011094_1 - 011094_5, 2021/03
In quasielastic neutron scattering studies, aluminum or aluminum alloys are frequently employed as sample cells. With the increasing incident-neutron flux, the research area currently continues to expand; thus, obtaining data has become quicker than ever for dilute conditions. One such area is the water-containing systems. In this study, we investigated the effect of temperature on Al and found that even in a low temperature atmosphere, Al corrosion can occur. This was attributed to the different thermal expansion coefficients of Al as a base substrate and Al oxide as a passivating film.
Tominaga, Taiki*; Kawakita, Yukinobu; Nakagawa, Hiroshi; Yamada, Takeshi*; Shibata, Kaoru
JPS Conference Proceedings (Internet), 33, p.011086_1 - 011086_5, 2021/03
We developed a quartz double cylindrical sample cell optimized for a backscattering neutron spectrometer, especially for BL02 (DNA), MLF in J-PARC. A quartz glass tube, with one end closed, is shaved to obtain a wall thickness of 0.55 mm. The inner tube is properly centered using a protrusion into the outer tube such that the interstice between the outer and inner tubes keeps constant. This quartz cell can be used for samples that should not be in contact with the aluminum surface. We verified cell's background effect between the quartz cell and Al cell by QENS measurements using D O buffer. The elastic intensity profiles of the buffer in a low Q region were identical between both quartz cell and Al cell (A1070). In a high Q region, however, the profiles were different caused by the first sharp diffraction peak of quartz glass. For this region the data should be analyzed by consideration of absorption correction and diffraction in individual thickness of quartz cell.
O buffer. The elastic intensity profiles of the buffer in a low Q region were identical between both quartz cell and Al cell (A1070). In a high Q region, however, the profiles were different caused by the first sharp diffraction peak of quartz glass. For this region the data should be analyzed by consideration of absorption correction and diffraction in individual thickness of quartz cell.
Inoue, Rintaro*; Oda, Takashi*; Nakagawa, Hiroshi; Tominaga, Taiki*; Saio, Tomohide*; Kawakita, Yukinobu; Shimizu, Masahiro*; Okuda, Aya*; Morishima, Ken*; Sato, Nobuhiro*; et al.
Scientific Reports (Internet), 10, p.21678_1 - 21678_10, 2020/12
Times Cited Count:8 Percentile:30.01(Multidisciplinary Sciences)Incoherent quasielastic neutron scattering (iQENS) is a fascinating technique for investigating the internal dynamics of protein. However, both low flux of neutron beam and absence of analytical procedure for extracting the internal dynamics from iQENS profile have been obstacles for studying it under physiological condition (in solution). Thanks to the recent development of neutron source, spectrometer and computational technique, they enable us to decouple internal dynamics, translational and rotational diffusions from the iQENS profile. The internal dynamics of two proteins: globular domain protein (GDP) and intrinsically disordered protein (IDP) in solution were studied. It was found that the average relaxation rate of IDP was larger than that of GDP. Through the detailed analyses on their internal dynamics, it was revealed that the fraction of mobile H atoms in IDP was much higher than that in GDP. Interestingly, the fraction of mobile H atoms was closely related to the fraction of H atoms on highly solvent exposed surfaces. The iQENS study presented that the internal dynamics were governed by the highly solvent exposed amino acid residues depending upon protein molecular architectures.
Nakano, Shota*; Konno, Soken*; Tomita, Naoto*; Matsuba, Go*; Tominaga, Taiki*; Takata, Shinichi
Microsystem Technologies, 22(1), p.57 - 63, 2016/01
Times Cited Count:3 Percentile:19.46(Engineering, Electrical & Electronic)We studied the structural formation process of polyacrylonitrile (PAN) gelation in a wide-spatial scale with the small and wide angle neutron scattering in MLF, J-PARC. In micron-scale structure, light transmittance and gelation time were evaluated. The presence of water in the mixed solvent accelerated the phase separation and gelation. In the opaque-gel case, we observed the nanometer-scaled correlation of crystalline cross-linking points and smooth boundary of two micron-scaled phases due to liquid-liquid phase separation. The correlation length is independent of the annealing time, then the cross-linking point exists locally and micro-gelation occurs during phase separation. By contrast, in the transparent-gel case, the correlation length was found to increase throughout the gelation process, and the entanglements of the polymer chains play a role similar to that of cross-linking points.
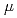 eV high energy resolution TOF type Si crystal analyzer backscattering spectrometer DNA at J-PARC/MLF
eV high energy resolution TOF type Si crystal analyzer backscattering spectrometer DNA at J-PARC/MLFShibata, Kaoru; Takahashi, Nobuaki*; Kawakita, Yukinobu; Matsuura, Masato*; Tominaga, Taiki*; Yamada, Takeshi*
no journal, ,
no abstracts in English
Endo, Hitoshi; Tominaga, Taiki; Takata, Shinichi; Matsumoto, Atsushi; Iwase, Hiroki*; Kamikubo, Hironari*; Kataoka, Mikio
no journal, ,
The dynamic and static structure factors for Staphylococcal Nuclease (SNase), which is a nucleolytic enzyme derived from Staphylococcus aureus, were evaluated by neutron spin echo (NSE)and small-angle neutron scattering (SANS) experiments. The SANS experiment was performed with TAIKAN (BL15) time-of-flight diffractometer at J-PARC/MLF, and we could obtain the static structure factors with wide Q range (0.2 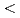 Q[1/
Q[1/![$AA] $<$$](/search/images/LRAUCXJAEQ4CI.gif) 2). The NSE measurement was performed with IN15 spectrometer at ILL. Grenoble, which enabled us to obtain intermediate scattering functions over 200 nanoseconds. The effects of hydration and internal motions were considered.
2). The NSE measurement was performed with IN15 spectrometer at ILL. Grenoble, which enabled us to obtain intermediate scattering functions over 200 nanoseconds. The effects of hydration and internal motions were considered.
Shibata, Kaoru; Kawakita, Yukinobu; Matsuura, Masato*; Tominaga, Taiki*; Yamada, Takeshi*
no journal, ,
no abstracts in English
Takata, Shinichi; Suzuki, Junichi*; Oishi, Kazuki*; Iwase, Hiroki*; Shinohara, Takenao; Oku, Takayuki; Nakatani, Takeshi; Inamura, Yasuhiro; Ito, Takayoshi*; Tominaga, Taiki*; et al.
no journal, ,
The small and wide angle neutron scattering instrument, 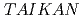 , was installed at the
, was installed at the 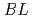 15 in the Materials and Life Science Experimental Facility,
15 in the Materials and Life Science Experimental Facility, 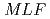 , of
, of 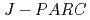 .
. 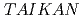 is designed for efficient measurement in wide-q range by using of the wide wavelength and the four detector banks which cover small-, middle-, high-, and backward-angle. At the present stage, 1,216
is designed for efficient measurement in wide-q range by using of the wide wavelength and the four detector banks which cover small-, middle-, high-, and backward-angle. At the present stage, 1,216  s are mounted on
s are mounted on  , where the number is about 50% of the installable total
, where the number is about 50% of the installable total  s number. On beam commissioning started in January 2012, and user program began in March 2012. The software of data reduction has newly developed and some sample environment devices such as a sample changer, tension tester, and refrigerator have been improved. In this presentation, we present the current status of
s number. On beam commissioning started in January 2012, and user program began in March 2012. The software of data reduction has newly developed and some sample environment devices such as a sample changer, tension tester, and refrigerator have been improved. In this presentation, we present the current status of  about the new software, sample environment devices, and the results on some samples to show the instrument performance of
about the new software, sample environment devices, and the results on some samples to show the instrument performance of  .
.
Tominaga, Taiki; Suzuki, Junichi*; Takata, Shinichi; Shinohara, Takenao; Oku, Takayuki; Nakatani, Takeshi; Inamura, Yasuhiro; Suzuya, Kentaro; Aizawa, Kazuya; Arai, Masatoshi; et al.
no journal, ,
no abstracts in English