Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Fujimoto, Hirofumi*; Higuchi, Mariko; Koike, Manabu*; Ode, Hirotaka*; Pinak, M.; Kotulic Bunta, J.*; Nemoto, Toshiyuki*; Sakudo, Takashi*; Honda, Naoko*; Maekawa, Hideaki*; et al.
Journal of Computational Chemistry, 33(3), p.239 - 246, 2012/01
Times Cited Count:36 Percentile:66.50(Chemistry, Multidisciplinary)Lysine acetylation is one of the most common protein post transcriptional modifications. The acetylation effects of lysine residues on Ku protein were examined herein applying several computer simulation techniques. Acetylation of the lysine residues did not reduce the affinity between Ku and its substrate, DNA, in spite of the fact that the substitution of lysine with glutamine (KQ mutant) reduced the affinity of Ku for DNA, or the substitution of lysine with arginine (KR mutant) did not reduce it, as previously reported in experimental studies. These results suggest that the effects of in vivo acetylation may be overestimated when the KQ mutant is employed in mimicry of the acetylated protein.
Yamauchi, Emiko*; Watanabe, Ritsuko; Oikawa, Miyoko*; Fujimoto, Hirofumi*; Yamada, Akinori*; Saito, Kimiaki; Murakami, Masahiro*; Hashido, Kazuo*; Tsuchida, Kozo*; Takada, Naoko*; et al.
Journal of Insect Biotechnology and Sericology, 77(1), p.17 - 24, 2008/02
The frequency of single strand breaks (SSBs) occurring on both strands of the pBR322 plasmid DNA region flanked by a pair of primers used for polymerase chain reaction (PCR) amplifications was determined after irradiation with 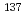 Cs
Cs  -rays. We refined that real time PCR is suitable for the detection and quantitative analysis of SSBs caused by
-rays. We refined that real time PCR is suitable for the detection and quantitative analysis of SSBs caused by  -ray irradiation. The utility of this approach was also supported by the comparison of the practical experimental data with the Monte Carlo simulation. The potential application of this PCR method for the detection of genomic DNA damage was also confirmed.
-ray irradiation. The utility of this approach was also supported by the comparison of the practical experimental data with the Monte Carlo simulation. The potential application of this PCR method for the detection of genomic DNA damage was also confirmed.