Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Hata, Kuniki; Urushibara, Ayumi*; Yamashita, Shinichi*; Lin, M.*; Muroya, Yusa*; Shikazono, Naoya; Yokoya, Akinari; Fu, H.*; Katsumura, Yosuke*
Journal of Radiation Research, 56(1), p.59 - 66, 2015/01
Times Cited Count:6 Percentile:27.54(Biology)Urushibara, Ayumi*; Kodama, Seiji*; Yokoya, Akinari
Mutation Research; Genetic Toxicology And Environmental Mutagenesis, 766, p.29 - 34, 2014/05
Times Cited Count:14 Percentile:41.73(Biotechnology & Applied Microbiology)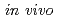 processing of clustered DNA damage
processing of clustered DNA damageShikazono, Naoya; Akamatsu, Ken; Takahashi, Momoko*; Noguchi, Miho; Urushibara, Ayumi; O'Neill, P.*; Yokoya, Akinari
Mutation Research; Fundamental and Molecular Mechanisms of Mutagenesis, 749(1-2), p.9 - 15, 2013/09
Times Cited Count:14 Percentile:39.16(Biotechnology & Applied Microbiology)We examined the biological consequences of bi-stranded clustered damage sites, consisting of a combination of DNA lesions using a bacterial plasmid-based assay. The transformation efficiencies were significantly lower for the bi-stranded clustered GAP/AP lesions than for either a single GAP or a single AP site. When the two lesions were separated by 10-20 bp, the transformation efficiencies were comparable with those of the single lesions. This recovery of transformation efficiency for separated lesions requires DNA polymerase I (Pol I) activity. Analogously, the mutation frequency was enhanced in a bi-stranded cluster containing a GAP and an 8-oxoG, and Pol I was found to play an important role in minimising mutations induced as a result of clustered lesions. These results indicate that the biological consequences of clustered DNA damage strongly depend on Pol I activity.
Hata, Kuniki; Urushibara, Ayumi; Yamashita, Shinichi; Shikazono, Naoya; Yokoya, Akinari; Katsumura, Yosuke*
Biochemical and Biophysical Research Communications, 434(2), p.341 - 345, 2013/05
Times Cited Count:8 Percentile:22.43(Biochemistry & Molecular Biology)Noguchi, Miho; Urushibara, Ayumi; Yokoya, Akinari; O'Neill, P.*; Shikazono, Naoya
Mutation Research; Fundamental and Molecular Mechanisms of Mutagenesis, 732(1-2), p.34 - 42, 2012/04
Times Cited Count:17 Percentile:45.22(Biotechnology & Applied Microbiology)The effect of a single strand break associated with base lesion(s) in vivo remains largely unknown. In the present study we determined the mutagenicities of two- and three-lesion clustered damage sites containing a 1-nucleotide gap (GAP) and 8-oxo-7,8-dihydroguanine(s) (8-oxoG(s)). The mutation frequencies (MFs) of bi-stranded two-lesion clusters (GAP/8-oxoG), especially in mutY-deficient strains, were high and were similar to those for bi-stranded clusters with 8-oxoG and base lesions/AP sites, suggesting that the GAP is processed with an efficiency similar to the efficiency of processing a base lesion or an AP site within a cluster. The MFs of tandem two-lesion clusters comprised of a GAP and an 8-oxoG were comparable to or less than the MF of a single 8-oxoG. The mutagenic potential of three-lesion clusters, which were comprised of a tandem lesion (a GAP and an 8-oxoG) and an opposing single 8-oxoG, was higher than that of a single 8-oxoG, but was no more than that of a bi-stranded 8-oxoGs. We suggest that incorporation of a nucleotide opposite 8-oxoG is less mutagenic when a GAP is present in a cluster than when a GAP is absent. Our observations indicate that the repair of a GAP is retarded by an opposing 8-oxoG, but not by a tandem 8-oxoG, and that the extent of GAP repair determines the biological consequences.
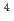 He
He ion irradiation
ion irradiationYokoya, Akinari; Shikazono, Naoya; Fujii, Kentaro; Noguchi, Miho; Urushibara, Ayumi
Radiation Protection Dosimetry, 143(2-4), p.219 - 225, 2011/02
Times Cited Count:3 Percentile:24.69(Environmental Sciences)Multiple single-strand breaks (m-SSBs), which are predicted to be preferentially induced by high LET radiation, would be underestimated if one uses the conventional method using plasmid DNA, because m-SSBs will not cause additionally conformational changes if they are on the same or on the opposite strand but separated each other sufficiently so as not to induce a double strand break. In order to observe the invisible m-SSBs, we have developed a novel technique using DNA denaturation. The m-SSBs arising in both strands of DNA are revealed as molecular size change in single strand DNA (SS-DNA) by gel electrophoresis. We have applied this method to the X- and He ion irradiated sample of hydrated pUC18 plasmid DNA. A half of SS-DNA population remains as intact within the experimental resolution (
ion irradiated sample of hydrated pUC18 plasmid DNA. A half of SS-DNA population remains as intact within the experimental resolution (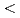 140 bases) for both irradiations. Contrary to our initial expectation, these results indicate that SSBs are not multiply induced over 140 bp even by high-LET irradiation.
140 bases) for both irradiations. Contrary to our initial expectation, these results indicate that SSBs are not multiply induced over 140 bp even by high-LET irradiation.
Shikazono, Naoya; Yokoya, Akinari; Urushibara, Ayumi; Noguchi, Miho; Fujii, Kentaro
Radiation Protection Dosimetry, 143(2-4), p.181 - 185, 2011/02
Times Cited Count:2 Percentile:17.52(Environmental Sciences)Shikazono, Naoya; Noguchi, Miho; Fujii, Kentaro; Urushibara, Ayumi*; Yokoya, Akinari
Journal of Radiation Research, 50(1), p.27 - 36, 2009/01
Times Cited Count:120 Percentile:88.18(Biology)After living cells are exposed to ionizing radiation, a variety of chemical modifications of DNA are induced either directly by ionization of DNA or indirectly through interactions with water-derived radicals. Clustered DNA damage, which is defined as two or more of such lesions within one to two helical turns of DNA induced by a single radiation track, is considered to be a unique feature of ionizing radiation. A double strand break (DSB) is a type of clustered DNA damage. Formation and repair of DSBs have been studied in great detail over the years as they have been linked to important biological endpoints. Although non-DSB clustered DNA damage has received less attention, there is growing evidence of its biological significance. This review focuses on the current understanding of (1) the yield of non-DSB clustered damage induced by ionizing radiation (2) the processing, and (3) biological consequences of non-DSB clustered DNA damage.
Yokoya, Akinari; Fujii, Kentaro; Shikazono, Naoya; Akamatsu, Ken; Urushibara, Ayumi; Watanabe, Ritsuko
International Journal of Radiation Biology, 84(12), p.1069 - 1081, 2008/12
Times Cited Count:12 Percentile:60.11(Biology)The role of Auger effect in inducing DNA damage has been studied using soft X-ray irradiation, which mainly cause photoelectric effect of DNA constituent atoms. As a consequence of Auger decay process, ejected low energy photo- or Auger-electrons might impact on proximately chemical group in the molecule. These highly localized collision events are expected to lead to a clustered DNA damage site within a few nano-meter. We have revealed that soft X-ray ( 60 keV) induced lesions visualized by the enzymatic probes show much higher yields than those induced by low LET
60 keV) induced lesions visualized by the enzymatic probes show much higher yields than those induced by low LET  -ray irradiation, and the yields decreased with decreasing soft X-ray energy (below a few keV). These results indicate that the complexity of damage site strongly depends on photo- or Auger electron range. The recent progress that has been made in the study of the process of DNA-radicals as precursors using an EPR apparatus combined with a synchrotron soft X-ray source is also presented.
-ray irradiation, and the yields decreased with decreasing soft X-ray energy (below a few keV). These results indicate that the complexity of damage site strongly depends on photo- or Auger electron range. The recent progress that has been made in the study of the process of DNA-radicals as precursors using an EPR apparatus combined with a synchrotron soft X-ray source is also presented.
Yokoya, Akinari; Shikazono, Naoya; Fujii, Kentaro; Ushigome, Takeshi*; Suzuki, Masao*; Urushibara, Ayumi; Watanabe, Ritsuko
Proceedings of the 27th Symposium on Materials Science and Engineering, Research Center of Ion Beam Technology Hosei University, 8 Pages, 2008/12
It has been indicated that ion particle irradiation to living cells causes a clustered DNA damage site, defined as two or more lesions formed within a few nano meters, in a cell nuclei. The clustered damage is less readily repaired by enzymatic repair system so that it induces biological effects, such as mutation induction. We have studied the mechanism of clustered damage induction by biochemical approaches using closed circular plasmid DNA as a model molecule to be irradiated with He ions from TIARA. We have used base excision repair proteins as enzymatic probes to quantify base lesions. We have also applied a novel method using DNA denaturation to visualize multiple single strand breaks which hardly be detected by conventional method. Our recent results will be reported in this seminar.
Yokoya, Akinari; Shikazono, Naoya; Fujii, Kentaro; Urushibara, Ayumi; Akamatsu, Ken; Watanabe, Ritsuko
Radiation Physics and Chemistry, 77(10-12), p.1280 - 1285, 2008/10
Times Cited Count:61 Percentile:95.60(Chemistry, Physical)Ionizing radiation induces a variety of damages in cellular DNA, which is thought to be the critical target of biological effects of radiation, by both direct energy deposition on DNA (direct effect) and reactions with diffusible water radicals (indirect effect). One of the goals of our study is to clarify the nature of DNA damage induced by direct effect. The yields of single- and double-strand breaks, base lesions and clustered damage induced in a plasmid DNA were measured after exposing to various kinds of radiation (ion particles; 20 to 500 keV/ 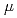 , photons; 0.4 keV to 1.3 MeV). Base excision repair enzymes were used to detect the oxidative base lesions. In order to obtain more detailed insights into the physicochemical mechanism of DNA damage induction, short-lived base radicals by applying an EPR spectrometer at a synchrotron ultrasoft X-ray beamline. Experimental evidences obtained by these methods will be discussed in comparison with the previous plasmid data.
, photons; 0.4 keV to 1.3 MeV). Base excision repair enzymes were used to detect the oxidative base lesions. In order to obtain more detailed insights into the physicochemical mechanism of DNA damage induction, short-lived base radicals by applying an EPR spectrometer at a synchrotron ultrasoft X-ray beamline. Experimental evidences obtained by these methods will be discussed in comparison with the previous plasmid data.
 He
He ion irradiation
ion irradiationUrushibara, Ayumi*; Shikazono, Naoya; O'Neill, P.*; Fujii, Kentaro; Wada, Seiichi*; Yokoya, Akinari
International Journal of Radiation Biology, 84(1), p.23 - 33, 2008/01
Times Cited Count:45 Percentile:92.11(Biology)To characterize the complexity of radiation damage to DNA, fully hydrated plasmid DNA was irradiated with 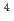 He
He ions. From quantification of the conformational changes of the irradiated samples, the yields of single-(SSB) and double strand break (DSB) were obtained. Base lesions were visualized as additional strand breaks by treatment with base excision repair enzymes. The yield of prompt SSBs does not depend significantly on LET of the
ions. From quantification of the conformational changes of the irradiated samples, the yields of single-(SSB) and double strand break (DSB) were obtained. Base lesions were visualized as additional strand breaks by treatment with base excision repair enzymes. The yield of prompt SSBs does not depend significantly on LET of the 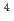 He
He ions, whereas the yield of prompt DSBs increases with increasing LET. The yields of isolated base lesions, revealed by enzymes as additional SSBs, decrease drastically with increasing LET. The sum of the yields of DSB and additional DSBs revealed by the enzymes increase with increasing LET of the
ions, whereas the yield of prompt DSBs increases with increasing LET. The yields of isolated base lesions, revealed by enzymes as additional SSBs, decrease drastically with increasing LET. The sum of the yields of DSB and additional DSBs revealed by the enzymes increase with increasing LET of the 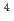 He
He ions except at the highest LET investigated. These results indicate that the yields of clustered damage, revealed as DSB and non-DSB clustered damage sites, increase with increasing ionization density of radiation.
ions except at the highest LET investigated. These results indicate that the yields of clustered damage, revealed as DSB and non-DSB clustered damage sites, increase with increasing ionization density of radiation.
Shikazono, Naoya; Urushibara, Ayumi; Fujii, Kentaro; Yokoya, Akinari
Hoshasen Seibutsu Kenkyu, 41(4), p.409 - 423, 2006/12
no abstracts in English
Yokoya, Akinari; Fujii, Kentaro; Ushigome, Takeshi; Shikazono, Naoya; Urushibara, Ayumi; Watanabe, Ritsuko
Radiation Protection Dosimetry, 122(1-4), p.86 - 88, 2006/12
Times Cited Count:13 Percentile:64.48(Environmental Sciences)We have studied yields of DNA damages induced by soft X-rays obtained from a conventional soft X-ray machine in a LET region between  -rays and ultrasoft X-rays. Practically soft X-rays with a broad energy spectrum emitted from a target of heavy metal, such as tungsten, have been widely used not only for radiobiological experiments but also for medical application such as mammography. Radiation weighting factors for these soft X-rays have been assigned to be 1 by ICRP. However, the fraction of a large number of low energy photons in the spectrum (below several tens keV) provided by bremsstrahlung is expected to be more effective for DNA damage induction than
-rays and ultrasoft X-rays. Practically soft X-rays with a broad energy spectrum emitted from a target of heavy metal, such as tungsten, have been widely used not only for radiobiological experiments but also for medical application such as mammography. Radiation weighting factors for these soft X-rays have been assigned to be 1 by ICRP. However, the fraction of a large number of low energy photons in the spectrum (below several tens keV) provided by bremsstrahlung is expected to be more effective for DNA damage induction than  -rays since low energy photo- and Auger electrons predominantly ejected in consequence of a photoelectric effect can produce dense clusters of ionization/excitation on DNA molecules. We have examined the yield of DNA strand breaks induced by white soft X-rays (150 kVp, tungsten target). Yields of base lesions revealed by base excision repair enzymes will be also presented.
-rays since low energy photo- and Auger electrons predominantly ejected in consequence of a photoelectric effect can produce dense clusters of ionization/excitation on DNA molecules. We have examined the yield of DNA strand breaks induced by white soft X-rays (150 kVp, tungsten target). Yields of base lesions revealed by base excision repair enzymes will be also presented.
Urushibara, Ayumi; Shikazono, Naoya; Watanabe, Ritsuko; Fujii, Kentaro; O'Neill, P.*; Yokoya, Akinari
Radiation Protection Dosimetry, 122(1-4), p.163 - 165, 2006/12
Times Cited Count:4 Percentile:29.72(Environmental Sciences)no abstracts in English
Yokoya, Akinari; Shikazono, Naoya; Urushibara, Ayumi; Fujii, Kentaro; Akamatsu, Ken; Watanabe, Ritsuko
Hoshasen Seibutsu Kenkyu, 40(2), p.168 - 184, 2005/06
Ionizing radiation causes modifications in a DNA molecule depending on the characteristic tack-structure in which two or more isolated lesions arise in a few nm scale (1 or 2 helical turn of DNA), known as "clustered DNA damage". These clustered DNA damages could be distinct from those by reactive oxygen species (ROS) endogenously induced on their severity of induction of biological effects such as mutation. However, the studies on the nature and repair mechanism of clustered DNA damage have still been behind because of the technical difficulties on determination of the chemical structure and yield. This article reviews some experimental evidences of the clustered DNA damages in this research field, as well as our recent progress on the studies on the clustered DNA damages using both molecular biological techniques and synchrotron spectroscopic method.
Yokoya, Akinari; Urushibara, Ayumi*; Fujii, Kentaro; Shikazono, Naoya
no journal, ,
One of the goals of our study is to clarify the nature of clustered damage induced by densely ionizing radiation. We have developed a novel technique using DNA denaturation by which irradiated DNA is analyzed as single strand DNA. The prompt or enzymatically revealed additional single strand breaks which arise in both strands of pUC18 plasmid DNA, but do not induce a double strand break are measured as degradation of single strand DNA using gel electrophoresis. To avoid induction of heat labile strand breaks by high temperature generally used for a denaturation treatment, we have determined much lower denaturation temperature (37 degrees centigrade) using formamide (50% v/v). Obtained preliminary results will be presented.
Noguchi, Miho; Urushibara, Ayumi*; Yokoya, Akinari; Shikazono, Naoya
no journal, ,
Clustered DNA damage is defined as two or more lesions induced within 1-2 helical turns (10-20bp) of DNA. Ionizing radiation-induced DNA damage, especially DNA double strand breaks (DSB) are considered to be a major factor for radiation induced chromosomal aberration and cell death. Many studies have shown the experimental evidence of DSB induction and its repair efficiency, as well as intracellular signal transduction caused by DSB. It is predicted that the non-DSB clustered damage shows high biological effects. It is, however, technically difficult to directly detect non-DSB clustered damage site as well as its effect in living cells. In this study, we investigated the potential of single strand break (SSB) to influence the mutagenicity of 8-oxoG in Escherichia coli. The mutation frequency of 8-oxoG was enhanced by the SSB situated on opposite strand. When SSB was present in tandem on the same strand of an 8-oxoG, however, these clustered damages showed lower mutation frequency than a single 8-oxoG lesion. These results suggest that SSB opposed to 8-oxoG have an inhibitory effect on repair of 8-oxoG, but that, in the case of 8-oxoG and SSB positioned in tandem, 8-oxoG can be removed, at least partly, by the repair process of SSB. We propose that the mutagenic potential of 8-oxoG depends on whether SSB is located on either strand, same or opposite, to 8-oxoG.
Yokoya, Akinari; Ushigome, Takeshi; Shikazono, Naoya; Fujii, Kentaro; Urushibara, Ayumi; Suzuki, Masao*; Tauchi, Hiroshi*; Watanabe, Ritsuko
no journal, ,
The yields of single- and double-strand breaks (SSB and DSB), base lesions and clustered damage induced in DNA were measured after exposing to various kinds of radiation. To focus on the effect of direct energy deposition from radiation track, we prepared hydrated DNA as well as solution sample with various scavenging capacities. Base excision repair enzymes, EndoIII and Fpg, were used to detect oxidative base lesions. The obtained results show that (1) the yield of directly induced SSB by the soft X-irradiation is about 30 percent of total SSBs in a cell mimetic condition and (2) the SSB yield does not significantly depend on the quality of radiation. On the other hand, (3) the yields of base lesions show a maximum by soft X-irradiation and drastically decreases with increasing of LET. (4) Soft X-rays are more effective in inducing base lesions than ions with similar LET, and (5) EndoIII treatment gives significantly higher SSB yield than those by Fpg treatment.
Noguchi, Miho; Urushibara, Ayumi*; Yokoya, Akinari; O'Neill, P.*; Shikazono, Naoya
no journal, ,
Ionizing radiation (IR) produces ionization and excitation along a radiation track, and generates, amongst other DNA lesions, clustered DNA damage sites in cellular DNA. Clustered DNA damage is defined as two or more lesions induced within 1-2 helical turns (10-20bp) of DNA. It has been reported that various types of SSB have a strong inhibitory effect on the excision of 8-oxo-7,8-dihydroguanine (8-oxoG) in vitro. In the present study, we used plasmid based assay in Escherichia coli to investigate the potential of SSB to influence the mutagenicity of 8-oxoG. As a model of clustered damage, we used synthesized oligonucleotides carrying an SSB and an 8-oxoG at a restriction enzyme recognition site. In single glycosylase-deficient strain mutY, the mutagenic potential of 8-oxoG was stimulated by an SSB situated on the opposite strand. This observation indicates that a SSB retards the incision of 8-oxoG by Fpg. On the other hand, when an SSB was placed in tandem with 8-oxoG on the same strand at varying separations, these tandem damage sites showed lower mutation frequency than a single 8-oxoG lesion in every strain. These results suggest that the 8-oxoG is removed, at least in part, during repair processing of the SSB. Our studies demonstrate that the mutagenic potential of 8-oxoG depends on whether the SSB is located on either the same strand or the opposite strand containing to 8-oxoG.