Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
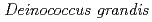 , isolated from freshwater fish in Japan
, isolated from freshwater fish in Japan佐藤 勝也; 小野寺 威文*; 面曽 宏太*; 武田-矢野 喜代子*; 片山 豪*; 大野 豊; 鳴海 一成*
Genome Announcements (Internet), 4(1), p.e01631-15_1 - e01631-15_2, 2016/01
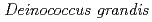 was isolated as a gram-negative, red-pigmented, radioresistant, rod-shaped bacterium from freshwater fish. The draft genome sequence of
was isolated as a gram-negative, red-pigmented, radioresistant, rod-shaped bacterium from freshwater fish. The draft genome sequence of 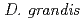 ATCC 43672 was 4,092,497 bp, with an average G+C content of 67.5% and comprised of 4 circular contigs and 3 linear contigs that was deposited at DDBJ/EMBL/Genbank under the accession number BCMS00000000. The draft genome sequence indicates that
ATCC 43672 was 4,092,497 bp, with an average G+C content of 67.5% and comprised of 4 circular contigs and 3 linear contigs that was deposited at DDBJ/EMBL/Genbank under the accession number BCMS00000000. The draft genome sequence indicates that 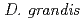 possesses a DNA damage response regulator (encoded by
possesses a DNA damage response regulator (encoded by 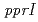 homolog) and radiation-desiccation response regulons (
homolog) and radiation-desiccation response regulons (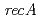 ,
, 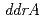 ,
, 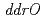 ,
, 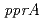 and
and 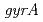 homologs etc.), which are involved in the unique radiation-desiccation response system in
homologs etc.), which are involved in the unique radiation-desiccation response system in 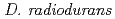 . As it is for
. As it is for 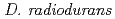 ,
, 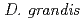 seems to employ the same radioresistant mechanisms. In future, the draft genome sequence of
seems to employ the same radioresistant mechanisms. In future, the draft genome sequence of 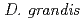 will be useful for elucidating the common principles of the radioresistance based on the extremely efficient DNA repair mechanisms in Deinococcus species by the comparative analysis of genomic sequences.
will be useful for elucidating the common principles of the radioresistance based on the extremely efficient DNA repair mechanisms in Deinococcus species by the comparative analysis of genomic sequences.
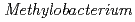 sp. ME121, isolated from soil as a mixed single colony with
sp. ME121, isolated from soil as a mixed single colony with 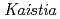 sp. 32K
sp. 32K藤浪 俊*; 武田 喜代子*; 小野寺 威文*; 佐藤 勝也; 清水 哲*; 若林 佑*; 鳴海 一成*; 中村 顕*; 伊藤 政博*
Genome Announcements (Internet), 3(5), p.e01005-15_1 - e01005-15_2, 2015/09
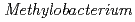 sp. ME121 was isolated from soil as a mixed single colony with
sp. ME121 was isolated from soil as a mixed single colony with 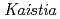 sp. 32K during our screening of L-glucose-utilizing microorganisms, and its growth was enhanced by coculture. It was expected that genomic analysis of this bacterium would provide novel information on coculture-dependent growth enhancement. The genomic information of symbiotic bacteria could be of use for studying the molecular mechanisms underlying microbial symbiosis. The draft genome sequence of
sp. 32K during our screening of L-glucose-utilizing microorganisms, and its growth was enhanced by coculture. It was expected that genomic analysis of this bacterium would provide novel information on coculture-dependent growth enhancement. The genomic information of symbiotic bacteria could be of use for studying the molecular mechanisms underlying microbial symbiosis. The draft genome sequence of 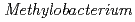 sp. ME121 is 7,096,979 bp in total length and comprises 197 large contigs (
sp. ME121 is 7,096,979 bp in total length and comprises 197 large contigs ( 500 bp) that was deposited at DDBJ/EMBL/GenBank under the accession number BBUX00000000. The draft genome sequence shows that
500 bp) that was deposited at DDBJ/EMBL/GenBank under the accession number BBUX00000000. The draft genome sequence shows that 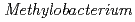 sp. ME121 has some genes that encode putative methanol/ethanol family PQQ-dependent dehydrogenases involved in methylotrophy. Some unknown factor provided by the coculture may contribute to increase the growth of
sp. ME121 has some genes that encode putative methanol/ethanol family PQQ-dependent dehydrogenases involved in methylotrophy. Some unknown factor provided by the coculture may contribute to increase the growth of 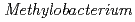 sp. ME121.
sp. ME121.
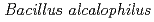 AV1934, a classic alkaliphile isolated from human feces in 1934
AV1934, a classic alkaliphile isolated from human feces in 1934Attie, O.*; Jayaprakash, A.*; Shah, H.*; Paulsen, I. T.*; 守野 正人*; 高橋 優嘉*; 鳴海 一成*; Sachidanandam, R.*; 佐藤 勝也; 伊藤 政博*; et al.
Genome Announcements (Internet), 2(6), p.e01175-14_1 - e01175-14_2, 2014/11
The alkaliphilic bacterium 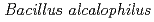 AV1934, isolated from human feces, recent has indicated novel potassium ion coupling to motility in this extremophile. We report draft sequences that will facilitate an examination of whether that coupling is part of a larger cycle of potassium ion-coupled transporters. This draft genome has 4,095 predicted genes and 4,063 predicted proteins. The whole-genome shotgun project has been deposited in Genbank under the accession no. ALPT02000000. In addition to the
AV1934, isolated from human feces, recent has indicated novel potassium ion coupling to motility in this extremophile. We report draft sequences that will facilitate an examination of whether that coupling is part of a larger cycle of potassium ion-coupled transporters. This draft genome has 4,095 predicted genes and 4,063 predicted proteins. The whole-genome shotgun project has been deposited in Genbank under the accession no. ALPT02000000. In addition to the 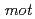 genes encoding the MotPS channel that uses either potassium or sodium coupling, two loci encoding multisubunit Mrp-type antiporters, were found. The
genes encoding the MotPS channel that uses either potassium or sodium coupling, two loci encoding multisubunit Mrp-type antiporters, were found. The 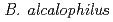 AV1934 genome reveals no tripartite ATP-independent (TRAP-T family) uptake systems, a major sodium-coupled complement in the other two alkaliphiles. The
AV1934 genome reveals no tripartite ATP-independent (TRAP-T family) uptake systems, a major sodium-coupled complement in the other two alkaliphiles. The 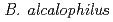 AV1934 genomic data will enable us to test the hypothesis of a major role of potassium in ion coupling in this extremophile.
AV1934 genomic data will enable us to test the hypothesis of a major role of potassium in ion coupling in this extremophile.
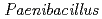 sp. strain TCA20, isolated from a hot spring containing a high concentration of calcium ions
sp. strain TCA20, isolated from a hot spring containing a high concentration of calcium ions藤浪 俊*; 武田 喜代子*; 小野寺 威文*; 佐藤 勝也; 佐野 元彦*; 高橋 優嘉*; 鳴海 一成*; 伊藤 政博*
Genome Announcements (Internet), 2(5), p.e00866-14_1 - e00866-14_2, 2014/09
Calcium-dependent  sp. strain TCA20 was isolated from a water sample of a hot spring containing a high concentration of calcium ions. Here, we report the draft genome sequence of this bacterium, which may be the basis for the research of calcium ion homeostasis. The draft genome sequence of
sp. strain TCA20 was isolated from a water sample of a hot spring containing a high concentration of calcium ions. Here, we report the draft genome sequence of this bacterium, which may be the basis for the research of calcium ion homeostasis. The draft genome sequence of  sp. strain TCA20 totals 5,631,463 bp in length and is composed of 33 large contigs (
sp. strain TCA20 totals 5,631,463 bp in length and is composed of 33 large contigs ( 500 bp) that was deposited at DDBJ/EMBL/Genbank under the accession no. BBIW00000000. The annotation of the draft genome sequence shows that
500 bp) that was deposited at DDBJ/EMBL/Genbank under the accession no. BBIW00000000. The annotation of the draft genome sequence shows that  sp. strain TCA20 has a calcium-specific calcium/proton antiporter, ChaA, and a P-type calcium-transporting ATPase, YloB, was identified as calcium transporters. These transporters may also be important for the growth of
sp. strain TCA20 has a calcium-specific calcium/proton antiporter, ChaA, and a P-type calcium-transporting ATPase, YloB, was identified as calcium transporters. These transporters may also be important for the growth of 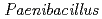 sp. strain TCA20 under the high concentration of calcium ions.
sp. strain TCA20 under the high concentration of calcium ions.
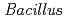 sp. strain TS-2, isolated from a jumping spider
sp. strain TS-2, isolated from a jumping spider藤浪 俊*; 武田 喜代子*; 小野寺 威文*; 佐藤 勝也; 佐野 元彦*; 鳴海 一成*; 伊藤 政博*
Genome Announcements (Internet), 2(3), p.e00458-14_1 - e00458-14_2, 2014/05
The potassium-dependent alkaliphilic 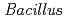 sp. strain TS-2 was isolated from the mashed extract of a jumping spider, and its draft genome sequence was obtained. Comparative genomic analysis with a previously sequenced sodium-dependent alkaliphilic
sp. strain TS-2 was isolated from the mashed extract of a jumping spider, and its draft genome sequence was obtained. Comparative genomic analysis with a previously sequenced sodium-dependent alkaliphilic 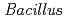 species may reveal potassium-dependent alkaline adaptation mechanisms. The draft genome sequence of
species may reveal potassium-dependent alkaline adaptation mechanisms. The draft genome sequence of 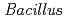 sp. strain TS-2 was 4,360,646 bp in total length, comprised 58 large contigs (
sp. strain TS-2 was 4,360,646 bp in total length, comprised 58 large contigs (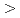 500 bp) that was deposited at DDBJ/EMBL/Genbank under the accession number BAWL00000000. In the draft genome sequence of
500 bp) that was deposited at DDBJ/EMBL/Genbank under the accession number BAWL00000000. In the draft genome sequence of 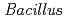 sp. strain TS-2, two sets of
sp. strain TS-2, two sets of 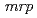 genes, which encode multisubunit secondary cation/proton antiporter-3 family proteins, were annotated. The Mrp complex acts as an Na
genes, which encode multisubunit secondary cation/proton antiporter-3 family proteins, were annotated. The Mrp complex acts as an Na /H
/H antiporter in typical alkaliphilic
antiporter in typical alkaliphilic 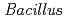 species and plays a critical role in the sodium-dependent alkaline adaptation mechanism.
species and plays a critical role in the sodium-dependent alkaline adaptation mechanism.
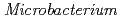 sp. strain TS-1
sp. strain TS-1藤浪 俊*; 武田 喜代子; 小野寺 威文*; 佐藤 勝也; 佐野 元彦*; 鳴海 一成*; 伊藤 政博*
Genome Announcements (Internet), 1(6), P. e01043-13, 2013/12
Alkaliphilic 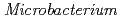 sp. strain TS-1, newly isolated from the jumping spider, showed Na
sp. strain TS-1, newly isolated from the jumping spider, showed Na -independent growth and motility. Here, we report the draft genome sequence of this bacterium, which may provide beneficial information for Na
-independent growth and motility. Here, we report the draft genome sequence of this bacterium, which may provide beneficial information for Na -independent alkaline adaptation mechanisms. The draft genome sequence of
-independent alkaline adaptation mechanisms. The draft genome sequence of 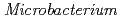 sp. strain TS-1 was 3,396,165 bp in total length and comprised 3 large contigs (2,699,404 bp, 691,278 bp, and 5,483 bp) that was deposited at DDBJ/ EMBL/GenBank under the accession number BASQ00000000. The annotation of the draft genome sequence indicated that this bacterium has 3 sets of
sp. strain TS-1 was 3,396,165 bp in total length and comprised 3 large contigs (2,699,404 bp, 691,278 bp, and 5,483 bp) that was deposited at DDBJ/ EMBL/GenBank under the accession number BASQ00000000. The annotation of the draft genome sequence indicated that this bacterium has 3 sets of 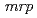 genes, which encode multisubunit secondary cation/proton antiporter-3 family proteins for pH homeostasis. The annotation also indicated that this bacterium has genes involved in chemotaxis/flagellar components, including one set of
genes, which encode multisubunit secondary cation/proton antiporter-3 family proteins for pH homeostasis. The annotation also indicated that this bacterium has genes involved in chemotaxis/flagellar components, including one set of 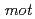 genes.
genes.