Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Fujimoto, Hirofumi*; Higuchi, Mariko; Koike, Manabu*; Ode, Hirotaka*; Pinak, M.; Kotulic Bunta, J.*; Nemoto, Toshiyuki*; Sakudo, Takashi*; Honda, Naoko*; Maekawa, Hideaki*; et al.
Journal of Computational Chemistry, 33(3), p.239 - 246, 2012/01
Times Cited Count:37 Percentile:64.85(Chemistry, Multidisciplinary)Lysine acetylation is one of the most common protein post transcriptional modifications. The acetylation effects of lysine residues on Ku protein were examined herein applying several computer simulation techniques. Acetylation of the lysine residues did not reduce the affinity between Ku and its substrate, DNA, in spite of the fact that the substitution of lysine with glutamine (KQ mutant) reduced the affinity of Ku for DNA, or the substitution of lysine with arginine (KR mutant) did not reduce it, as previously reported in experimental studies. These results suggest that the effects of in vivo acetylation may be overestimated when the KQ mutant is employed in mimicry of the acetylated protein.
Yamauchi, Emiko*; Watanabe, Ritsuko; Oikawa, Miyoko*; Fujimoto, Hirofumi*; Yamada, Akinori*; Saito, Kimiaki; Murakami, Masahiro*; Hashido, Kazuo*; Tsuchida, Kozo*; Takada, Naoko*; et al.
Journal of Insect Biotechnology and Sericology, 77(1), p.17 - 24, 2008/02
The frequency of single strand breaks (SSBs) occurring on both strands of the pBR322 plasmid DNA region flanked by a pair of primers used for polymerase chain reaction (PCR) amplifications was determined after irradiation with 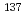 Cs
Cs  -rays. We refined that real time PCR is suitable for the detection and quantitative analysis of SSBs caused by
-rays. We refined that real time PCR is suitable for the detection and quantitative analysis of SSBs caused by  -ray irradiation. The utility of this approach was also supported by the comparison of the practical experimental data with the Monte Carlo simulation. The potential application of this PCR method for the detection of genomic DNA damage was also confirmed.
-ray irradiation. The utility of this approach was also supported by the comparison of the practical experimental data with the Monte Carlo simulation. The potential application of this PCR method for the detection of genomic DNA damage was also confirmed.
Fujimoto, Hirofumi*; Pinak, M.; Nemoto, Toshiyuki*; O'Neill, P.*; Kume, Etsuo; Saito, Kimiaki; Maekawa, Hideaki*
Journal of Computational Chemistry, 26(8), p.788 - 798, 2005/06
Times Cited Count:23 Percentile:57.53(Chemistry, Multidisciplinary)Clustered DNA damage sites induced by ionizing radiation have been suggested to have serious consequences to organisms. In this study, approaches based on molecular dynamics (MD) simulation have been applied to examine conformational changes and energetic properties of DNA molecules containing clustered damage sites consisting of 2 lesioned sites, 8-oxoG and AP site. After 1 nanosecond of MD simulation, one of the 6 DNA molecules containing a clustered damage site develop specific characteristic features: sharp bending at the lesioned site and weakening or complete loss of electrostatic interaction energy between 8-oxoG and bases locating on the complementary strand. From these results it is suggested that these changes would make it difficult for the repair enzyme to bind to the lesions within the clustered damage site and thereby result in a reduction of its repair capacity.
Pinak, M.; O'Neill, P.*; Fujimoto, Hirofumi; Nemoto, Toshiyuki*
AIP Conference Proceedings 708, p.310 - 313, 2004/05
The multiple nanosecond molecular dynamics simulations of DNA mutagenic oxidative lesion - 7,8-dihydro-8-oxoguanine (8-oxoG), complexed with the repair enzyme - human oxoguanine glycosylase 1 (hOGG1) in a physiological aqueous environment, were performed in order to describe the structural and energy changes in DNA and the dynamical process of DNA-enzyme complex formation. In complex the N-terminus of arginine 324 was found located close to the phosphodiester bond of the nucleotide with 8-oxoG enabling chemical reaction(s) between the amino acid and the lesion. The recognition of lesion on DNA, its recognition by repair enzyme and the formation of stable DNA-enzyme complex are necessary conditions for the onset of the successful enzymatic repair process.
Fujimoto, Hirofumi; Pinak, M.; Nemoto, Toshiyuki*; Sakamoto, Kiyotaka*; Yamada, Kazuyuki*; Hoshi, Yoshiyuki*; Kume, Etsuo
Journal of Molecular Structure; THEOCHEM, 681(1-3), p.1 - 8, 2004/01
We developed the novel system, Fujitsu Bio Molecular Visualization System (F-BMVS), that enables to produce real pictures and an animation by arranging them along a time series of a large scale simulation of biomolecules associated with a molecular dynamics (MD) simulation program. This animation system is used to study the results of molecular dynamics code, AMBER, in order to find structural differences on the lesioned DNA comparing with non-damaged DNA. These structural differences would be a factor that guides a repair enzyme to discriminate a lesion from non-damaged DNA region.