Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Kai, Takeshi; Yokoya, Akinari; Ukai, Masatoshi*; Fujii, Kentaro; Higuchi, Mariko; Watanabe, Ritsuko
Radiation Physics and Chemistry, 102, p.16 - 22, 2014/09
Times Cited Count:24 Percentile:86.35(Chemistry, Physical)no abstracts in English
Kai, Takeshi; Higuchi, Mariko; Fujii, Kentaro; Watanabe, Ritsuko; Yokoya, Akinari
International Journal of Radiation Biology, 88(12), p.928 - 932, 2012/12
Times Cited Count:1 Percentile:12.53(Biology)Fujimoto, Hirofumi*; Higuchi, Mariko; Koike, Manabu*; Ode, Hirotaka*; Pinak, M.; Kotulic Bunta, J.*; Nemoto, Toshiyuki*; Sakudo, Takashi*; Honda, Naoko*; Maekawa, Hideaki*; et al.
Journal of Computational Chemistry, 33(3), p.239 - 246, 2012/01
Times Cited Count:37 Percentile:64.85(Chemistry, Multidisciplinary)Lysine acetylation is one of the most common protein post transcriptional modifications. The acetylation effects of lysine residues on Ku protein were examined herein applying several computer simulation techniques. Acetylation of the lysine residues did not reduce the affinity between Ku and its substrate, DNA, in spite of the fact that the substitution of lysine with glutamine (KQ mutant) reduced the affinity of Ku for DNA, or the substitution of lysine with arginine (KR mutant) did not reduce it, as previously reported in experimental studies. These results suggest that the effects of in vivo acetylation may be overestimated when the KQ mutant is employed in mimicry of the acetylated protein.
Higuchi, Mariko; Fujii, Jumpei*; Yonetani, Yoshiteru; Kitao, Akio*; Go, Nobuhiro*
Journal of Structural Biology, 173(1), p.20 - 28, 2011/01
Times Cited Count:2 Percentile:6.15(Biochemistry & Molecular Biology)MutT distinguishes substrate 8-oxo-dGTP from dGTP and also 8-oxo-dGMP from dGMP despite small differences of chemical structures between them. In this paper we show by the method of molecular dynamics simulation that the transition between conformational substates of MutT is a key mechanism for a high resolution molecular recognition of the differences between the very similar chemical compounds. The native state MutT has two conformational substates with similar free energies, each characterized by either open or close of two loops surrounding the substrate binding active site. Between the two substates, the open substate is more stable in free MutT and in dGMP-MutT complex, and the closed substate is more stable in 8-oxo-dGMP-MutT complex. A hydrogen bond between H7 atom of 8-oxo-dGMP and the sidechain of Asn119 plays a crucial role for maintaining the closed substate in 8-oxo-dGMP-MutT complex.
Higuchi, Mariko; Pinak, M.; Saito, Kimiaki
Hoken Butsuri, 42(2), p.166 - 173, 2007/06
The cluster damaged site on DNA consisting of 7,8-dihydro-8-oxoguanine (8oxoG) and apurinic/apyrimidinic (AP) site is one example of damage that inhibits enzymatic repair. The 2 nanosecond molecular dynamics simulation revealed that the effect of AP site was lager than it of 8oxoG in multiple damaged DNA. The multiple damaged DNA was bent mainly at AP site. Bending direction of multiple damaged DNA including AP site and 8oxoG was different from that at the lesion site of single 8oxoG damaged DNA. These changes may have an influence on the effectiveness of enzymatic repair.
Ishida, Hisashi; Higuchi, Mariko; Yonetani, Yoshiteru*; Kano, Takuma; Jochi, Yasumasa*; Kitao, Akio*; Go, Nobuhiro
Annual Report of the Earth Simulator Center April 2005 - March 2006, p.237 - 240, 2007/01
no abstracts in English
Ishida, Hisashi; Higuchi, Mariko; Yonetani, Yoshiteru*; Kano, Takuma; Jochi, Yasumasa*; Kitao, Akio*; Go, Nobuhiro
Annual Report of the Earth Simulator Center April 2004 - March 2005, p.241 - 246, 2005/12
no abstracts in English
Ishida, Hisashi; Jochi, Yasumasa*; Higuchi, Mariko; Kano, Takuma; Kitao, Akio*; Go, Nobuhiro
Annual Report of the Earth Simulator Center April 2003 - March 2004, p.175 - 179, 2004/07
no abstracts in English
Higuchi, Mariko; Pinak, M.
no journal, ,
Ionizing radiation leads to the clustered DNA damage sites, which are defined as two or more single lesions induced within 10-20 base pairs (bp). The detail mechanism of repair of cluster lesions is not known. The experimental results showed that the excision of an 8-oxo-7,8-dihydroguanine 8-oxoG lesion by glycosylase MutM is strongly inhibited by the presence of an abasic (AP) site on the opposite strand with a 1bp separation from the 8-oxoG lesion. In this study, we present the results of molecular dynamics (MD) simulation, on comparison of the single 8-oxoG damaged DNA and the cluster damaged DNA containing AP site and 8-oxoG. In order to proceed with MD simulation we have calculated the molecular properties of damaged sites using quantum chemical approach. We have found the molecular structure and atomic charge distribution differ from those of non-damaged DNA parts.
Saito, Kimiaki; Watanabe, Ritsuko; Higuchi, Mariko; Ouchi, Noriyuki; Akamatsu, Ken; Kinase, Sakae
no journal, ,
no abstracts in English
Saito, Kimiaki; Watanabe, Ritsuko; Higuchi, Mariko
no journal, ,
no abstracts in English
Higuchi, Mariko; Pinak, M.; Saito, Kimiaki
no journal, ,
Ionizing radiation leads to clustered DNA damage sites. These sites are repaired with less efficiency than single lesions. So far the detail mechanism of repair of multiple lesions is not known. A lesion which locates within a few base pairs opposite to a 8oxo-G tends to inhibit excision of the 8oxo-G by repair enzyme MutM. The strength of inhibition effect is difference according to a kind of lesion. The inhibition effect of  -
-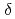 single strand break (SSB) is stronger than that of
single strand break (SSB) is stronger than that of  SSB and abasic (AP) site. In this study, we present the results of molecular dynamics (MD) simulation, by comparing (1)
SSB and abasic (AP) site. In this study, we present the results of molecular dynamics (MD) simulation, by comparing (1)  -
-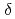 SSB, (2)
SSB, (2)  SSB and (3) the AP site in cluster damaged DNA which is containing 8-oxoG. We discuss with the relation between the structural change of cluster damaged DNA and the inhibition effect.
SSB and (3) the AP site in cluster damaged DNA which is containing 8-oxoG. We discuss with the relation between the structural change of cluster damaged DNA and the inhibition effect.
Higuchi, Mariko; Pinak, M.; Saito, Kimiaki
no journal, ,
Ionizing radiation leads to clustered DNA damage sites, which are defined as two or more single lesions induced within 10-20 base pairs (bp). The experimental results showed that the excision of the 8oxo-G lesion by repair enzyme hOGG1 is strongly inhibited by the presence of a SSB on the opposite strand with a 1bp separation from the 8oxo-G lesion. The detail mechanism of repair of multiple lesions is not known. In this study, we present the results of molecular dynamics simulation of hOGG1 and damaged DNA. We compare the result of single 8oxo-G DNA and hOGG1 complex and that of cluster damaged DNA. According to observed results, the structure of the DNA with SSB was stable with 8-oxoGs either flipped out or not. However in the DNA without SSB, the structure was stable only in the case of the 8-oxoG flip out.
Saito, Kimiaki; Higuchi, Mariko; Watanabe, Ritsuko
no journal, ,
Higuchi, Mariko; Pinak, M.
no journal, ,
Ionizing radiation leads to clustered DNA damage sites, which are defined as two or more single lesions induced within 10-20 base pairs (bp). In general, these sites are repaired with less efficiency than single lesions. So far the detail mechanism of repair of multiple lesions is not known. A cluster damage containing a single strand break (SSB) within a few base pairs opposite to a 8-oxo-7,8-dihydroguanine (8oxo-G) is one example of damage that is difficult to repair. In this study, we present results of molecular dynamics (MD) simulation of the models of complex of hOGG1 and the cluster damaged DNA. Each model DNA contains two damages as follows: (1) AP site and 8oxoG, (2) SSB and 8oxoG. The separation of the location of AP site and SSB from 8oxoG on the opposite strand were +1bp, -1bp and -3bp, respectively. According to the observed results, the structural deformation at the cluster depends on the relative positions of the AP site, or SSB, in respect to the 8-oxoG.
Higuchi, Mariko; Pinak, M.
no journal, ,
Ionizing radiation leads to clustered DNA damage sites, which are defined as two or more single lesions induced within 10-20 base pairs (bp). In general, these sites are repaired with less efficiency than single lesions. So far the detail mechanism of repair of multiple lesions is not known. A cluster damage containing a single strand break (SSB) within a few base pairs opposite to a 8-oxo-7,8-dihydroguanine (8oxoG) is one example of damage that is difficult to repair. In this study, we present results of molecular dynamics (MD) simulation of the models of complex of hOGG1 and cluster damaged DNA. Each model DNA contains two damages, SSB and 8oxoG. The separation of the location of the SSB from the 8oxoG on the opposite strand were +1bp, -1bp and -3bp, respectively. According to the observed results, the fluctuation of each damaged DNA was dependence on the separation between the two damages because the extent of contact with hOGG1 was different.
Higuchi, Mariko; Pinak, M.
no journal, ,
Ionizing radiation leads to clustered DNA damage sites, which are defined as two or more single lesions induced within 10-20 base pairs (bp). The detail mechanism of repair of multiple lesions is not known. The experimental results showed that the excision of the 8-oxo-7,8-dihydroguanine (8oxoG) lesion by repair enzyme hOGG1 is strongly inhibited by the presence of a single strand break (SSB) on the opposite strand with a 1bp separation from the 8oxoG lesion and probability of repair increases with increasing distance between the SSB and the 8oxoG. In this study, we present results of molecular dynamics simulation of the models of complex of hOGG1 and cluster damaged DNA. Each model DNA contains two damages, SSB and 8oxoG. According to the observed results, the structural fluctuation of cluster damaged DNA decreased largely when hOGG1 bound the cluster DNA. The effect of the SSB located at -3bp was small because the residue located at -3bp did not contact directly with hOGG1.
Higuchi, Mariko; Pinak, M.
no journal, ,
Ionizing radiation leads to clustered DNA damage sites, which are defined as two or more single lesions induced within 10-20 base pairs (bp). In general, these sites are repaired with less efficiency than single lesions. So far the detail mechanism of repair of multiple lesions is not known. A cluster damage containing a single strand break (SSB) within a few base pairs opposite to a 8-oxo-7,8-dihydroguanine (8oxoG) is one example of damage that is difficult to repair by hOGG1. In this study, we present results of molecular dynamics simulation of the models of complex of hOGG1 and cluster damaged DNA. Each model DNA contains SSB and 8oxoG. According to the estimated binding free energy, the affinity between hOGG1 and the DNA including SSB located -3bp from 8oxoG was smaller than that between hOGG1 and the DNA including SSB located +3bp from 8oxoG. This result supported the relation between inhibition effects by cluster damaged DNA and affinity between damaged DNA and hOGG1.
Higuchi, Mariko; Pinak, M.
no journal, ,
Ionizing radiation leads to clustered DNA damage sites, which are defined as two or more single lesions induced within 10-20 base pairs (bp). In general, these sites are repaired with less efficiency than single lesions. So far the detail mechanism of repair of multiple lesions is not known. A cluster damage containing a single strand break (SSB) within a few base pairs opposite to a 8-oxo-7,8-dihydroguanine (8oxoG) is one example of damage that is difficult to repair by hOGG1. In this study, we present results of molecular dynamics simulation of the complex of hOGG1 and cluster damaged DNA. We prepared the several damaged DNA models which are varied relative location between the SSB and the 8oxoG. According to the estimated binding free energy, the affinity between hOGG1 and the damaged DNA was qualitatively compatible with the probability of excision of 8oxoG by hOGG1. This result supported the relation between inhibition effects by cluster damaged DNA.
Kai, Takeshi; Higuchi, Mariko; Fujii, Kentaro; Watanabe, Ritsuko; Yokoya, Akinari
no journal, ,
no abstracts in English