Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Savage, D.*; Benbow, S.*; Watson, C.*; Takase, Hiroyasu*; Ono, Kaori*; Oda, Chie; Honda, Akira
Applied Clay Science, 47(1-2), p.72 - 81, 2010/01
Times Cited Count:37 Percentile:70.43(Chemistry, Physical)Mudstones containing smectite have been altered under mildly alkaline conditions (9 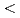 pH
pH 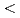 10) at Searles Lake, California over a 3 million-year time period. This natural alteration has been simulated incorporating time-dependent boundary conditions of sedimentation and fluid composition, a Pitzer model for activities of aqueous species, and a coupled hydrogeological model for time-dependent flow in the sediment layers. Kinetic dissolution of detrital smectite under alkaline conditions was described using one of two models based on departure from thermodynamic equilibrium or by an empirical rate dependent upon aqueous Si concentrations. The zonal pattern of smectite dissolution observed at Searles Lake was reproduced reasonably well by the "Cama-TST" model of montmorillonite dissolution. This assessment provides a test of the accuracy and reliability of published data in the application of models of smectite dissolution in the long-term.
10) at Searles Lake, California over a 3 million-year time period. This natural alteration has been simulated incorporating time-dependent boundary conditions of sedimentation and fluid composition, a Pitzer model for activities of aqueous species, and a coupled hydrogeological model for time-dependent flow in the sediment layers. Kinetic dissolution of detrital smectite under alkaline conditions was described using one of two models based on departure from thermodynamic equilibrium or by an empirical rate dependent upon aqueous Si concentrations. The zonal pattern of smectite dissolution observed at Searles Lake was reproduced reasonably well by the "Cama-TST" model of montmorillonite dissolution. This assessment provides a test of the accuracy and reliability of published data in the application of models of smectite dissolution in the long-term.
Yamasaki, Chisato*; Murakami, Katsuhiko*; Fujii, Yasuyuki*; Sato, Yoshiharu*; Harada, Erimi*; Takeda, Junichi*; Taniya, Takayuki*; Sakate, Ryuichi*; Kikugawa, Shingo*; Shimada, Makoto*; et al.
Nucleic Acids Research, 36(Database), p.D793 - D799, 2008/01
Times Cited Count:51 Percentile:71.25(Biochemistry & Molecular Biology)Here we report the new features and improvements in our latest release of the H-Invitational Database, a comprehensive annotation resource for human genes and transcripts. H-InvDB, originally developed as an integrated database of the human transcriptome based on extensive annotation of large sets of fulllength cDNA (FLcDNA) clones, now provides annotation for 120 558 human mRNAs extracted from the International Nucleotide Sequence Databases (INSD), in addition to 54 978 human FLcDNAs, in the latest release H-InvDB. We mapped those human transcripts onto the human genome sequences (NCBI build 36.1) and determined 34 699 human gene clusters, which could define 34 057 protein-coding and 642 non-protein-coding loci; 858 transcribed loci overlapped with predicted pseudogenes.