Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
石田 恒; Hayward, S.*
Biophysical Journal, 95(12), p.5962 - 5973, 2008/12
被引用回数:29 パーセンタイル:60.5(Biophysics)Molecular dynamics simulations were performed on  70S ribosome with and without the nascent polypeptide inside the exit tunnel. Modeling of the polypeptide in the tunnel revealed two possible paths: one over Arg
70S ribosome with and without the nascent polypeptide inside the exit tunnel. Modeling of the polypeptide in the tunnel revealed two possible paths: one over Arg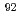 of L22 and one under (from the viewpoint of 50S on top of 30S). A strong interaction between L4 and Arg
of L22 and one under (from the viewpoint of 50S on top of 30S). A strong interaction between L4 and Arg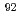 was observed without the polypeptide and when it passed over Arg
was observed without the polypeptide and when it passed over Arg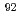 . However, when the polypeptide passed under, Arg
. However, when the polypeptide passed under, Arg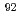 repositioned to interact with Ade
repositioned to interact with Ade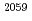 of 23S rRNA. Using steered molecular dynamics the polypeptide could be pulled through the L4-L22 constriction when situated under Arg
of 23S rRNA. Using steered molecular dynamics the polypeptide could be pulled through the L4-L22 constriction when situated under Arg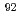 , but did not move when over. These results suggest that the tunnel is closed by the Arg
, but did not move when over. These results suggest that the tunnel is closed by the Arg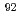 -L4 interaction before elongation of the polypeptide and the tunnel leads the entering polypeptide from the peptidyl transferase center to the passage under Arg
-L4 interaction before elongation of the polypeptide and the tunnel leads the entering polypeptide from the peptidyl transferase center to the passage under Arg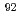 , causing Arg
, causing Arg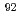 to switch to an open position. It is possible, therefore, that Arg
to switch to an open position. It is possible, therefore, that Arg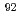 plays the role of a gate, opening and closing the tunnel at L4-L22. There is controversy over whether the tunnel is dynamics or rigid. At least within the time-scale of our simulations conformational analysis showed that global motions mainly involve relative movement of the 50S and 30S subunits and appear not to affect the conformation of the tunnel.
plays the role of a gate, opening and closing the tunnel at L4-L22. There is controversy over whether the tunnel is dynamics or rigid. At least within the time-scale of our simulations conformational analysis showed that global motions mainly involve relative movement of the 50S and 30S subunits and appear not to affect the conformation of the tunnel.
Poornam, G. P.*; 松本 淳; 石田 恒; Hayward, S.*
Proteins: Structure, Function, and Bioinformatics, 76(1), p.201 - 212, 2008/12
被引用回数:83 パーセンタイル:91.05(Biochemistry & Molecular Biology)A new method for the analysis of domain movements in large, multichain, biomolecular complexes is presented. The method is applicable to any molecule for which two atomic structures are available that represent a conformational change indicating a possible domain movement. The method is blind to atomic bonding and atom type and can, therefore, be applied to biomolecular complexes containing different constituent molecules such as protein, RNA, or DNA. Here, we report on the application of the method to biomolecules covering a considerable size range: hemoglobin, liver alcohol dehydrogenase, S-Adenosylhomocysteine hydrolase, aspartate transcarbamylase, and the 70S ribosome. The results provide a depiction of the conformational change within each molecule that is easily understood, giving a perspective that is expected to lead to new insights.