Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Hirata, Yoshinobu*; Nakagawa, Hiroshi; Yamauchi, Hiroki; Kaneko, Koji; Hagihara, Masato; Yamaguchi, Hideyuki*; Imaizumi, Teppei*; Nishizu, Takahisa*
International Journal of Biological Macromolecules, 306, p.141668_1 - 141668_7, 2025/03
Times Cited Count:0 Percentile:0.00(Biochemistry & Molecular Biology)We used quasi-elastic neutron scattering (QENS) to investigate the effect of the ratio of water to rice on the molecular dynamics of cooked rice starch during retrogradation. X-ray diffraction and differential scanning calorimetric measurements revealed that the degree of recrystallization and change in enthalpy were smaller with an increase in the amount of water added for cooking rice, whereas little difference in the crystallinity of the gelatinized rice starch was detected. The QENS measurements determined that the elastic incoherent structure factor (EISF) values of gelatinized samples were smaller with an increase in the amount of water added for cooking rice, indicating that the molecular dynamics of gelatinized rice starch with higher added water content were spatially more extended.
Inoue, Rintaro*; Oda, Takashi; Nakagawa, Hiroshi; Tominaga, Taiki*; Ikegami, Takahisa*; Konuma, Tsuyoshi*; Iwase, Hiroki*; Kawakita, Yukinobu; Sato, Mamoru*; Sugiyama, Masaaki*
Biophysical Journal, 124(3), p.540 - 548, 2025/02
Times Cited Count:0 Percentile:0.00(Biophysics)Hirata, Yoshinobu*; Kaneko, Fumitoshi*; Radulescu, A.*; Nishizu, Takahisa*; Katsuno, Nakako*; Imaizumi, Teppei*; Motokawa, Ryuhei; Kumada, Takayuki; Nakagawa, Hiroshi
Journal of Applied Glycoscience, 72(1), p.7201102_1 - 7201102_9, 2025/02
A simultaneous measurement system of SANS/FTIR-ATR was applied to record multiple structural changes in potato starch during retrogradation. In the SANS patterns, the shoulder-like peak becomes more pronounced with time. The peak intensity, Imax, which represents the amount of orderly arranged double helical structures, increased over time, showing that starch reassembled orderly on the nanoscale upon retrogradation. In the FTIR-ATR spectra, the increase of the ratio of absorptions at 1042 cm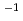 and 1016 cm
and 1016 cm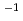 , indicating the amount of short-range ordered structure in starch increased during retrogradation. The time to obtain half of the equilibrium value for
, indicating the amount of short-range ordered structure in starch increased during retrogradation. The time to obtain half of the equilibrium value for 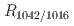 was larger than that for Imax. Changes in the short-range ordered structure of starch were observed to converge prior to changes in the nanostructure. These results indicate that the double-helix structures are initially formed by amylopectin side chains, and then these double-helical structures are arranged orderly.
was larger than that for Imax. Changes in the short-range ordered structure of starch were observed to converge prior to changes in the nanostructure. These results indicate that the double-helix structures are initially formed by amylopectin side chains, and then these double-helical structures are arranged orderly.
Kawai, Kiyoshi*; Sogabe, Tomochika*; Nakagawa, Hiroshi; Yamada, Takeshi*; Koseki, Shigenobu*
Journal of Food Engineering, 375, p.112066_1 - 112066_9, 2024/08
Times Cited Count:1 Percentile:31.11(Engineering, Chemical)The purpose of this study was to clarify effect of glycerol and glucose on the mechanical glass transition and dynamical transition of freeze-dried bacteria at various water activity (aw)-conditions. From the water sorption isotherm, it was noted that the water content at each aw and monolayer water content were higher in the order of glycerol, glucose, non-added samples. Effect of temperature on the mean square displacement (MSD) of atoms in the samples was investigated by incoherent elastic neutron scattering. The MSD increased gradually with an increase in temperature depending on the aw and added solute. From the linear fitting, three dynamical transition temperatures (low-, middle- and high-Tds) were determined. The Couchman-Karasz model suggested that the added solute and bacteria were not completely miscible.
Kikuchi, Shin; Kondo, Toshiki; Doi, Daisuke; Seino, Hiroshi; Ogawa, Kengo*; Nakagawa, Takeshi*
Proceedings of 14th International Topical Meeting on Nuclear Reactor Thermal-Hydraulics, Operation, and Safety (NTHOS-14) (Internet), 12 Pages, 2024/08
Murmiliuk, A.*; Iwase, Hiroki*; Kang, J.-J.*; Mohanakumar, S.*; Appavou, M.-S.*; Wood, K.*; Alm sy, L.*; Len, A.*; Schw
sy, L.*; Len, A.*; Schw rzer, K.*; Allgaier, J.*; et al.
rzer, K.*; Allgaier, J.*; et al.
Journal of Colloid and Interface Science, 665, p.801 - 813, 2024/07
Times Cited Count:6 Percentile:74.56(Chemistry, Physical)The complexity of protein structure limits our ability to predict and tune the properties of the formed nanoparticles. The goal of our research is to elucidate the key triggers of the morphological transition in protein/PE complexes, evaluate their encapsulation efficacy, and assess particle stability by the systematic study of complexes formed by block copolymers with proteins and ionic drugs. We demonstrated that copolymers consisting of PE and neutral hydrophilic block co-assemble with insulin at pH values close to the protein isoelectric point. The insulin arrangement within the particle is controlled by electrostatic forces between protein molecules, and the morphology of the formed particles can be tuned by varying pH and ionic strength.
Nakagawa, Hiroshi; Hirata, Yoshinobu*; Nishizu, Takahisa*
Teion Seibutsu Kogakkai-Shi, 70(1), p.17 - 24, 2024/06
Inelastic neutron scattering can measure the atomic thermal fluctuations in pico- to nano-second time scales, and is the powerful method to observe the low-energy dynamics in THz region and to characterize the motional geometry of bio-molecules. It is widely used in material science research, such as the study of the molecular motion of water using the strong scattering from hydrogen atoms and the molecular motion of glass materials, but it is also effective as a probe to directly observe the molecular motion of various biomolecules such as proteins and starch, and food molecules. Here, the characteristics of neutron beams and the fundamentals of inelastic neutron scattering spectra will be explained. Then, three typical examples of studies on the obtained molecular motions will be presented. These research examples may be applicable to various issues in cryo-biology.
Kumada, Takayuki; Nakagawa, Hiroshi; Miura, Daisuke; Sekine, Yurina; Motokawa, Ryuhei; Hiroi, Kosuke; Inamura, Yasuhiro; Oku, Takayuki; Oishi, Kazuki*; Morikawa, Toshiaki*; et al.
Hamon, 34(2), p.50 - 53, 2024/05
Spin-contrast-variation (SCV) small-angle neutron scattering (SANS) enabled us to determine structure of nano-ice crystals that were generated in rapidly frozen sugar solution. In the frozen glucose solution, we found that the nano-ice crystals formed a planar structure with a radius larger than several tens of nanometers and a thickness of 2-3 nm, which was close to the critical nucleation size of ice crystals in supercooled water. This result suggests that the glucose molecules were preferentially bound to a specific face of nano-ice crystals, and then blocked the crystal growth perpendicular to that face.
Muhialdin, B. J.*; Sanchez, C. F.*; Nakagawa, Hiroshi; Ubbink, J.*
Food Biophysics, 19(1), p.172 - 181, 2024/03
Times Cited Count:2 Percentile:19.17(Food Science & Technology)The impact of molecular interactions on the physical properties of extruded pea protein isolate (PPI) is investigated by adding interaction-modulating compounds to the matrix premix and studying the resulting variations in mechanical and physicochemical properties. Blends of PPI containing either sodium phosphate, urea, sodium dodecylsulphate and beta-mercaptoethanol, as well as with all four compounds and only with deionized water were extruded into thin strands using a lab-scale twin-screw extruder. The hardness from texture profile analysis was the lowest for matrices extruded with beta-mercaptoethanol and with all four chemicals, and highest for the control sample. The water holding capacity of the matrices is lowest for the beta-mercaptoethanol-containing matrix. Our results confirm the importance of disulfide bonds in texturized PPI and show that hydrophobic and electrostatic interactions play auxiliary roles in modulating the properties of extruded PPI matrices.
Kumada, Takayuki; Motokawa, Ryuhei; Oba, Yojiro; Nakagawa, Hiroshi; Sekine, Yurina; Micheau, C.; Ueda, Yuki; Sugita, Tsuyoshi; Birumachi, Atsushi; Sasaki, Miki; et al.
Journal of Applied Crystallography, 56(6), p.1776 - 1783, 2023/12
Times Cited Count:18 Percentile:97.57(Chemistry, Multidisciplinary)The combination of the existing position-sensitive photomultiplier and the  He main detector with focusing devices, and the newly installed front detectors in SANS-J at JRR-3 covers small-angle neutron scattering signals in the range of the magnitude of the scattering vector Q from 0.002 to 6 nm-1 gaplessly with three standard device layouts. The installation of the front detector and a graphical user interface system largely improved the usability of SANS-J.
He main detector with focusing devices, and the newly installed front detectors in SANS-J at JRR-3 covers small-angle neutron scattering signals in the range of the magnitude of the scattering vector Q from 0.002 to 6 nm-1 gaplessly with three standard device layouts. The installation of the front detector and a graphical user interface system largely improved the usability of SANS-J.
Kutsukake, Kenichi; Matsuda, Makoto; Nakamura, Masahiko; Ishizaki, Nobuhiro; Kabumoto, Hiroshi; Otokawa, Yoshinori; Asozu, Takuhiro; Matsui, Yutaka; Nakagawa, Sohei; Abe, Shinichi
Proceedings of 20th Annual Meeting of Particle Accelerator Society of Japan (Internet), p.1080 - 1084, 2023/11
no abstracts in English
Kaneko, Fumitoshi*; Radulescu, A.*; Nakagawa, Hiroshi
Journal of Applied Crystallography, 56(5), p.1522 - 1527, 2023/10
Times Cited Count:2 Percentile:41.77(Chemistry, Multidisciplinary)Small angle neutron scattering (SANS) is widely used as a powerful technique to study the higher-order structure of soft matter. To increase the reliability of SANS profile analysis for complex, multi-component systems, combining different types of structural information obtained by other methods is desirable. A simultaneous measurement system combining SANS and Fourier transform infrared (FTIR) spectroscopy meets this objective and is particularly useful for targets for which it is difficult to match the timing of structural changes between experiments, but there is a problem that samples suitable for SANS are too thick for typical transmission FTIR measurements. To overcome this problem, a new simultaneous measurement system that employs the attenuated total reflection sampling method for FTIR spectroscopy has been developed.
Hirata, Yoshinobu*; Nakagawa, Hiroshi; Yamauchi, Hiroki; Kaneko, Koji; Hagihara, Masato; Yamaguchi, Hideyuki*; Omoto, Chie*; Katsuno, Nakako*; Imaizumi, Teppei*; Nishizu, Takahisa*
Food Hydrocolloids, 141, p.108728_1 - 108728_7, 2023/08
Times Cited Count:13 Percentile:79.03(Chemistry, Applied)Crystallinity is reflected in the mechanical properties of foods and materials. Crystallinity should be related to the structural dynamics of starch. In this study, we used quasi-elastic neutron scattering (QENS) to investigate changes in the molecular dynamics of cooked rice starch during retrogradation. The width of the measured QENS narrowed with retrogradation. The elastic incoherent structure factor (EISF) increased, which indicated that the molecular dynamics are spatially suppressed upon retrogradation. Analysis of EISF with a bimodal continuous diffusion model, where low and high mobilities are assumed to correspond to crystalline and amorphous phases, respectively, showed that the fraction of the low-mobility component increases with retrogradation.
Kumada, Takayuki; Nakagawa, Hiroshi; Miura, Daisuke; Sekine, Yurina; Motokawa, Ryuhei; Hiroi, Kosuke; Inamura, Yasuhiro; Oku, Takayuki; Oishi, Kazuki*; Morikawa, Toshiaki*; et al.
Journal of Physical Chemistry Letters (Internet), 14(34), p.7638 - 7643, 2023/08
Times Cited Count:0 Percentile:0.00(Chemistry, Physical)The structure of nano-ice crystals in rapidly frozen glucose solution was elucidated by using spin-contrast-variation small-angle neutron scattering, which distinguishes the nano-ice crystal signal from the frozen amorphous solution signal by the polarization-dependent neutron scattering. The analysis revealed that the nano-ice crystals form a planar structure with a diameter exceeding tens of nanometers and a thickness of 1 nm, which is close to the critical nucleation size. This result suggests that the glucose molecules are preferentially bound to a specific face of nano-ice crystals, and then block the crystal growth perpendicular to that face.
Kabumoto, Hiroshi; Nakagawa, Sohei; Matsuda, Makoto
JAEA-Conf 2022-002, 146 Pages, 2023/03
"The 34th Meeting for Tandem Accelerators and their Associated Technologies" was held on July 21-22, 2022 organized by Nuclear Science Research Institute of the Japan Atomic Energy Agency. This meeting was held only on-line for preventing the spread of COVID-19 infection. The purpose of this meeting is contribution of development for related technology and of management of facilities through exchange of information among the researchers and engineers using and operating electrostatics accelerator facilities like tandem accelerators. There were 25 presentations which contains current status report of facility, technical development of accelerator, research of application. The total number of participants was a hundred, from 26 universities, research organizations and industries. This meeting consisted of only oral session, a poster session was not carried out because of on-line meeting. This proceeding compiles the contents of report papers in the meeting.
Nakagawa, Hiroshi; Yamamoto, Naoki*
Life (Internet), 13(2), p.318_1 - 318_15, 2023/02
Times Cited Count:2 Percentile:14.65(Biology)Incoherent neutron scattering and terahertz spectroscopy have approximately the same energy range of measurement. Since both techniques are used to study the dynamics of proteins and hydrated water, it is important to review the advantages and disadvantages of both techniques and the relevant literature. To the best of our knowledge, there is no review of both methods, and we believe that this review is of high value.
Kabumoto, Hiroshi; Matsuda, Makoto; Nakamura, Masahiko; Ishizaki, Nobuhiro; Kutsukake, Kenichi; Otokawa, Yoshinori; Asozu, Takuhiro; Matsui, Yutaka; Nakagawa, Sohei; Abe, Shinichi
Proceedings of 19th Annual Meeting of Particle Accelerator Society of Japan (Internet), p.1109 - 1113, 2023/01
no abstracts in English
Sogabe, Tomochika*; Nakagawa, Hiroshi; Yamada, Takeshi*; Koseki, Shigenobu*; Kawai, Kiyoshi*
Biophysical Journal, 121(20), p.3874 - 3882, 2022/10
Times Cited Count:3 Percentile:27.83(Biophysics)The purpose of this study was to clarify the glass transition behavior of bacteria (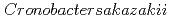 ) as a function of water activity (
) as a function of water activity (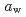 ). Mechanical relaxation was investigated at 298 K, and the mechanical
). Mechanical relaxation was investigated at 298 K, and the mechanical 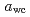 (
(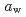 at which mechanical glass transition occurs at 298 K) was determined to be 0.667. Temperature-dependency of mean square displacement was investigated by inelastic neutron scattering. From the linear fitting, two dynamical transition temperatures (low and high-
at which mechanical glass transition occurs at 298 K) was determined to be 0.667. Temperature-dependency of mean square displacement was investigated by inelastic neutron scattering. From the linear fitting, two dynamical transition temperatures (low and high-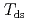 ) were determined. There was a minor effect of
) were determined. There was a minor effect of 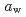 on the low-
on the low-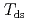 except for the anhydrous sample. The high-
except for the anhydrous sample. The high-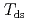 largely increased with the decrease in
largely increased with the decrease in 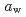 . The dynamical
. The dynamical 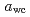 determined by high-
determined by high-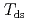 (0.688) was slightly higher than the mechanical
(0.688) was slightly higher than the mechanical 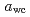 because of the difference in the measurement time-scale. The high-
because of the difference in the measurement time-scale. The high-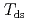 was converted to the glass transition temperature (
was converted to the glass transition temperature (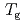 ), and anhydrous
), and anhydrous 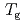 was estimated to be 411 K. Bacterial inactive-active transition was discussed according to the glass transition behavior.
was estimated to be 411 K. Bacterial inactive-active transition was discussed according to the glass transition behavior.
Nakagawa, Hiroshi; Matsuo, Tatsuhito*
Furontoranna Jikiden Sobunri Kaiseki Purotokoru; Jikken Igaku Bessatsu Saikyo No Suteppu UP Shirizu, p.222 - 227, 2022/07
Although the principle of neutron scattering is the same as that of X-ray scattering, it provides unique information due to the different interaction with the sample during the scattering process. Small-angle neutron scattering and quasi-elastic neutron scattering are promising methods to analyze the structure and association of proteins in droplets and their molecular dynamics. Neutron scattering experiments, which require nuclear reactors or accelerators as sources, are limited in Japan, and overseas facilities may be used if necessary.
Tominaga, Taiki*; Nakagawa, Hiroshi; Sahara, Masae*; Oda, Takashi*; Inoue, Rintaro*; Sugiyama, Masaaki*
Life (Internet), 12(5), p.675_1 - 675_9, 2022/05
Times Cited Count:2 Percentile:20.29(Biology)The background scattering of sample cells suitable for aqueous protein solution samples, conducted with a neutron backscattering spectrometer, was evaluated. It was found that the scattering intensity of an aluminum sample cell coated with boehmite using D O was lower than that of a sample cell coated with regular water (H
O was lower than that of a sample cell coated with regular water (H O). In addition, meticulous attention to cells with small individual weight differences and the positional reproducibility of the sample cell relative to the spectrometer neutron beam position enabled the accurate subtraction of the scattering profiles of the D
O). In addition, meticulous attention to cells with small individual weight differences and the positional reproducibility of the sample cell relative to the spectrometer neutron beam position enabled the accurate subtraction of the scattering profiles of the D O buffer and the sample container. Consequently, high quality information on protein dynamics could be extracted from dilute protein solutions.
O buffer and the sample container. Consequently, high quality information on protein dynamics could be extracted from dilute protein solutions.