Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Ishitsuka, Etsuo; Ho, H. Q.; Kitagawa, Kanta*; Fukuda, Takahito*; Ito, Ryo*; Nemoto, Masaya*; Kusunoki, Hayato*; Nomura, Takuro*; Nagase, Sota*; Hashimoto, Haruki*; et al.
JAEA-Technology 2023-013, 19 Pages, 2023/06
Eight people from five universities participated in the 2022 summer holiday practical training with the theme of "Technical development on HTTR". The participants practiced the feasibility study for nuclear battery, the burn-up analysis of HTTR core, the feasibility study for 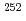 Cf production, the analysis of behavior on loss of forced cooling test, and the thermal-hydraulic analysis near reactor pressure vessel. In the questionnaire after this training, there were impressions such as that it was useful as a work experience, that some students found it useful for their own research, and that discussion with other university students was a good experience. These impressions suggest that this training was generally evaluated as good.
Cf production, the analysis of behavior on loss of forced cooling test, and the thermal-hydraulic analysis near reactor pressure vessel. In the questionnaire after this training, there were impressions such as that it was useful as a work experience, that some students found it useful for their own research, and that discussion with other university students was a good experience. These impressions suggest that this training was generally evaluated as good.
Yamasaki, Chisato*; Murakami, Katsuhiko*; Fujii, Yasuyuki*; Sato, Yoshiharu*; Harada, Erimi*; Takeda, Junichi*; Taniya, Takayuki*; Sakate, Ryuichi*; Kikugawa, Shingo*; Shimada, Makoto*; et al.
Nucleic Acids Research, 36(Database), p.D793 - D799, 2008/01
Times Cited Count:52 Percentile:71.15(Biochemistry & Molecular Biology)Here we report the new features and improvements in our latest release of the H-Invitational Database, a comprehensive annotation resource for human genes and transcripts. H-InvDB, originally developed as an integrated database of the human transcriptome based on extensive annotation of large sets of fulllength cDNA (FLcDNA) clones, now provides annotation for 120 558 human mRNAs extracted from the International Nucleotide Sequence Databases (INSD), in addition to 54 978 human FLcDNAs, in the latest release H-InvDB. We mapped those human transcripts onto the human genome sequences (NCBI build 36.1) and determined 34 699 human gene clusters, which could define 34 057 protein-coding and 642 non-protein-coding loci; 858 transcribed loci overlapped with predicted pseudogenes.
Watanabe, Tomoaki; Kikuchi, Takeo; Kasuya, Yuta*; Nomura, Takuro*; Suyama, Kenya
no journal, ,
JAEA has developed and maintained a burnup calculation code system SWAT4 to evaluate the nuclide composition of spent fuel. Due to the validations, various post-irradiation examinations (PIE) analyses have been performed using the JENDL-4.0 cross-section data. In this study, we performed the previous PIE analyses using the newly released JENDL-5 cross section data to confirm the changes in the analysis results due to the change to JENDL-5 and the differences in the results due to the reactor system and fuel type of the targeted PIE data. As a result of the analysis, it was found that the C/E values of nuclide composition by JENDL-5 did not show any significant difference in trend by reactor system or fuel type, and the results of JENDL-5 and JENDL-4.0 were similar in accuracy for all reactor systems and fuel types. As for the effect of the updated cross sections from JENDL-4.0 to JENDL-5 on C/E values, significant changes in C/E values for Pu-238, Am-241, and Cs-134 were observed due to the updated capture cross sections for Pu-238, Am-241, and Cs-133, independent of reactor system or fuel type existed. The results were highly dependent on the PIE samples as to whether the differences from the measured values were improved or not.