Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Okazaki, Nobuo*; Hattori, Takanori
CROSS Reports (Internet), 3, p.001_1 - 001_8, 2025/02
There have been requests for remote analysis of measured data at the BL11 PLANET beamline in the Materials and Life Science Experimental Facility (MLF) of the Japan Proton Accelerator Research Complex (J-PARC), but a system to perform this analysis on a regular basis has not been established. In order to meet this demand, a system for remote analysis was established using NoMachine, which is widely used as a remote desktop connection environment. This system is built on the cloud, and users can analyze data from anywhere with an Internet environment by using the NoMachine client.
Okazaki, Nobuo*; Hattori, Takanori
CROSS Reports (Internet), 1, p.001_1 - 001_9, 2023/12
There has been no method for users to monitor the current status of the experiments at BL11 (PLANET) from outside. To solve this, a monitoring system, named RemoShot, has been implemented to capture remotely the screenshots of the control PC. The RemoShot consists of a screenshot control server, a control PC in the beamline, and cloud storage. It operates in the following sequence: 1) Start a screenshot control server on the PC at beamline, 2) The screenshot control server accesses the MLF-side PC (measurement control PC) via SSH authentication and capturesa screenshot, 3) the captured screenshot is stored in cloud storage, and 4) after authentication, users can see the screenshot in the cloud. This system allows users to check the status of their experiments from anywhere using smartphones and other devices, and to quickly revise the experimental plan in case of measurement issuers or sequence delays due to unexpected beam off. We also discussed security and operating costs of this system.
Hattori, Takanori; Sano, Asami; Machida, Shinichi*; Abe, Jun*; Funakoshi, Kenichi*; Arima, Hiroshi*; Okazaki, Nobuo*
High Pressure Research, 39(3), p.417 - 425, 2019/06
Times Cited Count:29 Percentile:81.55(Physics, Multidisciplinary)We have developed a technique for neutron diffraction experiments at pressures up to 40 GPa using a Paris-Edinburgh press at the PLANET beamline in J-PARC. To increase the maximum accessible pressure, the diameter of the dimple for sample chamber at the top of the sintered diamond anvils is sequentially reduced from 4.0 mm to 1.0 mm. As a result, the maximum pressure increased and finally reached 40 GPa. By combining this technique with the beam optics which defines the gauge volume, diffraction patterns sufficient for full-structure refinements are obtainable at such pressures.
Hattori, Takanori; Sano, Asami; Machida, Shinichi*; Abe, Jun*; Funakoshi, Kenichi*; Okazaki, Nobuo*
Nihon Kessho Gakkai-Shi, 59(6), p.301 - 308, 2017/12
PLANET is a neutron beamline dedicated to high-pressure experiments. Combining the intense neutron source of J-PARC and high-pressure devices designed for time-of-flight powder neutron diffraction enables precise structure analysis of crystal, liquid and amorphous solids over wide pressure and temperature region of 0-20 GPa and 77-2000K. This beamline is effective for various studies in geophysics, planetary science, physics and chemistry. This paper overviews the beamline and introduces recent results obtained at PLANET.
Sakasai, Kaoru; Sato, Setsuo*; Seya, Tomohiro*; Nakamura, Tatsuya; To, Kentaro; Yamagishi, Hideshi*; Soyama, Kazuhiko; Yamazaki, Dai; Maruyama, Ryuji; Oku, Takayuki; et al.
Quantum Beam Science (Internet), 1(2), p.10_1 - 10_35, 2017/09
Neutron devices such as neutron detectors, optical devices including supermirror devices and  He neutron spin filters, and choppers are successfully developed and installed at the Materials Life Science Facility (MLF) of the Japan Proton Accelerator Research Complex (J-PARC), Tokai, Japan. Four software components of MLF computational environment, instrument control, data acquisition, data analysis, and a database, have been developed and equipped at MLF. MLF also provides a wide variety of sample environment options including high and low temperatures, high magnetic fields, and high pressures. This paper describes the current status of neutron devices, computational and sample environments at MLF.
He neutron spin filters, and choppers are successfully developed and installed at the Materials Life Science Facility (MLF) of the Japan Proton Accelerator Research Complex (J-PARC), Tokai, Japan. Four software components of MLF computational environment, instrument control, data acquisition, data analysis, and a database, have been developed and equipped at MLF. MLF also provides a wide variety of sample environment options including high and low temperatures, high magnetic fields, and high pressures. This paper describes the current status of neutron devices, computational and sample environments at MLF.
Hattori, Takanori; Sano, Asami; Arima, Hiroshi*; Funakoshi, Kenichi*; Abe, Jun*; Machida, Shinichi*; Okazaki, Nobuo*; Ouchi, Keiichi*; Inamura, Yasuhiro
Koatsuryoku No Kagaku To Gijutsu, 26(2), p.89 - 98, 2016/06
PLANET is a high-pressure neutron beamline constructed at pulsed-neutron source in Materials and Life Science Facility (MLF) in J-PARC. The six-axis multi-anvil press designed for time of flight (TOF) neutron diffraction experiments enables routine data collection at high pressures and high temperatures up to 10 GPa and 2000 K, respectively. To obtain clean data, the beamline is equipped with the incident slits and receiving collimators that eliminate parasitic scattering from the high-pressure cell. The high performance of the diffractometer for the resolution (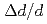
 0.6%) and the accessible d-spacing range (0.2 - 8.4
0.6%) and the accessible d-spacing range (0.2 - 8.4  ) together with low-parasitic scattering characteristics enables precise structure determination of crystals and liquids under high pressure and temperature conditions.
) together with low-parasitic scattering characteristics enables precise structure determination of crystals and liquids under high pressure and temperature conditions.
 -lactamase from the moderate halophile
-lactamase from the moderate halophile  sp.560 and the discovery of a Cs
sp.560 and the discovery of a Cs -selective binding site
-selective binding siteArai, Shigeki; Yonezawa, Yasushi*; Okazaki, Nobuo*; Matsumoto, Fumiko*; Shibazaki, Chie; Shimizu, Rumi; Yamada, Mitsugu*; Adachi, Motoyasu; Tamada, Taro; Kawamoto, Masahide*; et al.
Acta Crystallographica Section D, 71(3), p.541 - 554, 2015/03
Times Cited Count:8 Percentile:52.42(Biochemical Research Methods)The crystal structure of halophilic  -lactamase from
-lactamase from  sp.560 (HaBLA) was determined using X-ray crystallography. Moreover, the locations of bound Sr
sp.560 (HaBLA) was determined using X-ray crystallography. Moreover, the locations of bound Sr and Cs
and Cs ions were identified by anomalous X-ray diffraction. The location of one Cs
ions were identified by anomalous X-ray diffraction. The location of one Cs specific binding site was identified on HaBLA even in the presence of 9-fold molar excess of Na
specific binding site was identified on HaBLA even in the presence of 9-fold molar excess of Na (90 mM Na
(90 mM Na /10 mM Cs
/10 mM Cs ). This Cs
). This Cs binding site is formed by two main-chain O atoms and an aromatic ring of a side chain of Trp. An aromatic ring of Trp interacts with Cs
binding site is formed by two main-chain O atoms and an aromatic ring of a side chain of Trp. An aromatic ring of Trp interacts with Cs by the cation-
by the cation-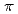 interaction. The observation of a selective and high-affinity Cs
interaction. The observation of a selective and high-affinity Cs binding site provides important information that is useful for designing artificial Cs
binding site provides important information that is useful for designing artificial Cs binding sites useful in bioremediation of radioactive isotopes.
binding sites useful in bioremediation of radioactive isotopes.
Okazaki, Hiro; Sumi, Mika; Sato, Mitsuhiro; Kayano, Masashi; Kageyama, Tomio; Martinez, P.*; Xu, N.*; Thomas, M.*; Porterfield, D.*; Colletti, L.*; et al.
Kaku Busshitsu Kanri Gakkai (INMM) Nihon Shibu Dai-35-Kai Nenji Taikai Rombunshu (Internet), 9 Pages, 2015/01
The quality control section of Plutonium Fuel Development Center (PFDC) in Japan Atomic Energy Agency has been analyzing isotopic compositions and content of plutonium and uranium as well as impurity and physics of nuclear materials in the process of MOX fuel fabrication for accountancy purpose as well as process control purposes. These analytical techniques are also effective for nuclear forensics to identify the source, history, and route of the material by determining a composition and chemical property of it. Therefore, PFDC cooperates with Los Alamos National Laboratory which has broad experience and established measurement skill for nuclear forensics, and evaluates the each method, procedure, and analytical data toward R&D of characterizing a nuclear fuel for forensics purposes. This paper describes the approaches to develop characterization techniques of nuclear fuel for nuclear forensic purpose at PFDC.
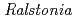 sp. A-471 reveals a unique arrangement of the catalytic residues for inverting chitin hydrolysis
sp. A-471 reveals a unique arrangement of the catalytic residues for inverting chitin hydrolysisArimori, Takao*; Kawamoto, Noriko*; Shinya, Shoko*; Okazaki, Nobuo*; Nakazawa, Masami*; Miyatake, Kazutaka*; Fukamizo, Tamo*; Ueda, Mitsuhiro*; Tamada, Taro
Journal of Biological Chemistry, 288(26), p.18696 - 18706, 2013/07
Times Cited Count:31 Percentile:62.77(Biochemistry & Molecular Biology)Chitinase C from 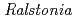 sp. A-471 (Ra-ChiC) has a catalytic domain sequence similar to goose type (G-type) lysozymes and, unlike other chitinases, belongs to glycohydrolase (GH) family 23. Using NMR spectroscopy, however, Ra-ChiC was found to interact only with the chitin dimer but not with the peptideglycan fragment. Here we report the crystal structures of wild-type, E141Q, and E162Q of the catalytic domain of Ra-ChiC with or without chitin oligosaccharides. Ra-ChiC has a substrate-binding site including a tunnel-shaped cavity, which determines the substrate specificity. Mutation analyses based on this structural information indicated that a highly conserved Glu141 acts as a catalytic acid, and that Asp226 located at the roof of the tunnel activates a water molecule as a catalytic base. The unique arrangement of the catalytic residues makes a clear contrast to the other GH23 members and also to inverting GH19 chitinases.
sp. A-471 (Ra-ChiC) has a catalytic domain sequence similar to goose type (G-type) lysozymes and, unlike other chitinases, belongs to glycohydrolase (GH) family 23. Using NMR spectroscopy, however, Ra-ChiC was found to interact only with the chitin dimer but not with the peptideglycan fragment. Here we report the crystal structures of wild-type, E141Q, and E162Q of the catalytic domain of Ra-ChiC with or without chitin oligosaccharides. Ra-ChiC has a substrate-binding site including a tunnel-shaped cavity, which determines the substrate specificity. Mutation analyses based on this structural information indicated that a highly conserved Glu141 acts as a catalytic acid, and that Asp226 located at the roof of the tunnel activates a water molecule as a catalytic base. The unique arrangement of the catalytic residues makes a clear contrast to the other GH23 members and also to inverting GH19 chitinases.
 KM1
KM1Okazaki, Nobuo; Tamada, Taro; Feese, M. D.*; Kato, Masaru*; Miura, Yutaka*; Komeda, Toshihiro*; Kobayashi, Kazuo*; Kondo, Keiji*; Blaber, M.*; Kuroki, Ryota
Protein Science, 21(4), p.539 - 552, 2012/04
Times Cited Count:5 Percentile:10.88(Biochemistry & Molecular Biology) sp. nucleoside diphosphate kinase
sp. nucleoside diphosphate kinaseArai, Shigeki; Yonezawa, Yasushi; Okazaki, Nobuo; Matsumoto, Fumiko; Tamada, Taro; Tokunaga, Hiroko*; Ishibashi, Matsujiro*; Blaber, M.; Tokunaga, Masao*; Kuroki, Ryota
Protein Science, 21(4), p.498 - 510, 2012/04
Times Cited Count:15 Percentile:34.81(Biochemistry & Molecular Biology)In order to clarify the oligomer state of nucleoside diphosphate kinase (NDK) from moderately halophilic  sp. 593 (HaNDK), the crystal structure of HaNDK was determined by X-ray crystallography. The crystal structures of the wild-type HaNDK and the mutant HaNDK (E134A) showed a dimer and a tetramer, respectively. The higher ordered association of proteins usually contributes to an increase in thermal stability and substrate affinity. The change in the assembly form by a minimum mutation may be an effective way for NDK to acquire molecular characteristics suited to various circumstances.
sp. 593 (HaNDK), the crystal structure of HaNDK was determined by X-ray crystallography. The crystal structures of the wild-type HaNDK and the mutant HaNDK (E134A) showed a dimer and a tetramer, respectively. The higher ordered association of proteins usually contributes to an increase in thermal stability and substrate affinity. The change in the assembly form by a minimum mutation may be an effective way for NDK to acquire molecular characteristics suited to various circumstances.
 HB8
HB8Okazaki, Nobuo; Adachi, Motoyasu; Tamada, Taro; Kurihara, Kazuo; Oga, Takushi*; Kamiya, Nobuo*; Kuramitsu, Seiki*; Kuroki, Ryota
Acta Crystallographica Section F, 68(1), p.49 - 52, 2012/01
Times Cited Count:2 Percentile:31.75(Biochemical Research Methods) sp. A-471
sp. A-471Okazaki, Nobuo; Arimori, Takao; Nakazawa, Masami*; Miyatake, Kazutaka*; Ueda, Mitsuhiro*; Tamada, Taro
Acta Crystallographica Section F, 67(4), p.494 - 497, 2011/04
Times Cited Count:3 Percentile:40.30(Biochemical Research Methods)Kuroki, Ryota; Okazaki, Nobuo; Adachi, Motoyasu; Ohara, Takashi; Kurihara, Kazuo; Tamada, Taro
Acta Crystallographica Section D, 66(11), p.1126 - 1130, 2010/11
Times Cited Count:2 Percentile:29.19(Biochemical Research Methods)It is generally known that enzymes represent important drug-target proteins. Elucidation of the catalytic function and the molecular-recognition mechanisms of enzymes provides important information for structure-based drug design. Neutron crystallography provides accurate information on the locations of H atoms that are essential in enzymatic function and molecular recognition. Recent examples are described of the structure determination of the drug-target proteins human immunodeficiency virus protease and porcine pancreatic elastase in complex with transition-state analogue inhibitors using the neutron diffractometers for biological crystallography (BIX-3 and BIX-4) installed at the JRR-3 research reactor.
Kurihara, Kazuo; Okazaki, Nobuo; Kuroki, Ryota
Radioisotopes, 59(4), p.263 - 277, 2010/04
Crystallographic analysis using neutron diffraction allows identification and position determination of light atoms like hydrogen. This method has been used for three-dimensional structure determination of organic compounds as well as macromolecules like protein. Hydrogen atoms in proteins, as well as those in solvent molecules, play significant roles in many naturally occurring processes, such as catalytic function and molecular recognition. In the field of neutron crystallography novel diffractometers and techniques for preparation and crystallization of target samples has been developed to complement the low flux of neutron sources to permit higher measurement performance. In Japan, single-crystal diffractometers named BIX-3 and BIX-4 were constructed with Neutron Imaging Plates as a detector. These diffractometers have contributed to the investigation of hydrogen-related molecular structures; for example, determination of hydrogen atom positions which are difficult to predict based on X-ray structure data, precise configuration of hydrogen bonds, and the orientation degree of freedom of hydration water molecules. On the other hand, a complementary application of neutron diffraction with X-ray diffraction has also been developed. Using a joint structure refinement method with X-ray diffraction data, elucidation of an enzymatic reaction mechanism and observation of a particular atomic configuration including hydrogen atoms were successfully achieved in neutron crystallographic studies of drug-discovery-target proteins. The information obtained from these neutron analyses has been consolidated into a database called Hydrogen and Hydration Database for Biomolecules which permits the analysis of key statistical information. In Japan as well as overseas, efforts to acquire higher measurement performance are now in progress to further investigate mechanisms involving hydrogen atoms, and to increase the application of neutron crystallographic studies.
Adachi, Motoyasu; Ohara, Takashi; Kurihara, Kazuo; Tamada, Taro; Honjo, Eijiro; Okazaki, Nobuo; Arai, Shigeki; Shoyama, Yoshinari; Kimura, Kaname*; Matsumura, Hiroyoshi*; et al.
Proceedings of the National Academy of Sciences of the United States of America, 106(12), p.4641 - 4646, 2009/03
Times Cited Count:114 Percentile:90.42(Multidisciplinary Sciences)To further understand the catalytic mechanism and inhibitor recognition of HIV-1 protease, we need to determine the locations of key hydrogen atoms in the catalytic aspartates Asp25 and Asp125. The structure of HIV-1 protease in complex with transition-state analog KNI-272 was determined by combined neutron crystallography at 1.9  resolution and X-ray crystallography at 1.4
resolution and X-ray crystallography at 1.4  resolution. The resulting structural data shows that the catalytic residue Asp25 is protonated and that Asp125 is deprotonated. The proton on Asp25 makes a hydrogen bond with the carbonyl group of the allophenylnorstatine group in KNI-272. The deprotonated Asp125 bonds to the hydroxyl proton of Apns. The results provide direct experimental evidence for proposed aspects of the catalytic mechanism of HIV-1 protease; and can therefore contribute substantially to the development of specific inhibitors for therapeutic application.
resolution. The resulting structural data shows that the catalytic residue Asp25 is protonated and that Asp125 is deprotonated. The proton on Asp25 makes a hydrogen bond with the carbonyl group of the allophenylnorstatine group in KNI-272. The deprotonated Asp125 bonds to the hydroxyl proton of Apns. The results provide direct experimental evidence for proposed aspects of the catalytic mechanism of HIV-1 protease; and can therefore contribute substantially to the development of specific inhibitors for therapeutic application.
Okazaki, Nobuo; Ohara, Takashi; Umino, Hisao*; Chatake, Toshiyuki*; Kurihara, Kazuo; Cachau, R. E.*; Blaber, M.*; Niimura, Nobuo*; Kuroki, Ryota
no journal, ,
In protein molecules, key energetic contributors are solvation, desolvation and hydrogen bonding. They contribute protein folding, dynamics and molecular recognition. As a result, more elaborate studies of hydrogen atoms will be great help to recognize protein structures and obtain new findings of them. However, we do not have system which dedicated to characterization and analysis of hydrogen bonding. Therefore, we have developed a database for hydrogen and hydration water molecules. That database named Hydrogen and Hydration Database for Biomolecules (HHDB). Hydrogen bond data stored to HHDB use hydrogen atom coordinates determined directly by neutron diffraction and certain extremely high resolution X-ray diffraction. HHDB provides graphical user interface, users can use it through web browser. HHDB can visualize hydrogen atom positions in protein and solvent, and hydrogen bonding interactions. Fig. 1 shows HHDB plot example. In this plot, hydrogen atom is placed at the origin, and each point represents hydrogen bond distance and angle. We are improving the web user interfaces and the performance for usability.
Arai, Shigeki; Yonezawa, Yasushi; Okazaki, Nobuo; Tamada, Taro; Tokunaga, Hiroko*; Ishibashi, Matsujiro*; Tokunaga, Masao*; Kuroki, Ryota
no journal, ,
The nucleoside diphosphate kinases (NDKs) are known to have a tetrameric or hexameric oligomer structure formed by association of common dimeric components. We determined the crystal structure of E134A mutant NDK from Halomonas sp. 593 (HaNDK) and found that two kinds of tetrameric assemblies, Type I seen in the Myxococcus NDK tetramer and Type II seen in the E.coli NDK tetramer, appeared in the asymmetric unit. Change in the assembly form may be an effective way for NDK to acquire molecular characteristics suited to various circumstances.
Tamada, Taro; Kurihara, Kazuo; Ohara, Takashi; Okazaki, Nobuo; Kuroki, Ryota
no journal, ,
Neutron crystallography, which is a powerful technique for locating hydrogen atoms, enables us to obtain accurate atomic positions within proteins. Neutron diffraction data can provide information of the location of hydrogen atoms to the structural information determined by X-ray crystallography. There are two neutron diffractometers for biological crystallography (BIX-3 and BIX-4) installed at research reactor (JRR-3) in Japan Atomic Energy Agency (JAEA), which have been contributed to 15 structure analyses of biological macromolecules. Here, we report the recent activities for developments of the neutron diffractometers and the structure analysis of drug-target proteins.
 sp.593 (HaNDK)
sp.593 (HaNDK)Arai, Shigeki; Yonezawa, Yasushi; Okazaki, Nobuo; Tamada, Taro; Tokunaga, Hiroko*; Ishibashi, Matsujiro*; Tokunaga, Masao*; Kuroki, Ryota
no journal, ,
The nucleoside diphosphate kinases (NDKs) are known to have a tetrameric or hexameric oligomer structure formed by association of common dimeric components. In order to understand the oligomeric structure of NDK, the wild-type NDK from Halomonas sp. 593 (HaNDK) was determined by X-ray crystallography. The wild-type HaNDK only exhibited a dimeric form that is the same as a common dimer unit seen in other NDKs. This is the first evidence of a dimeric structure for HaNDK. To explore the effect of mutation on the assembly form of HaNDK, E134, which is a key interface residue for other tetrameric NDKs, was replaced with Ala, and the structure was determined by X-ray crystallography. Two kinds of tetrameric assemblies, Type I and Type II, appeared in the asymmetric unit of the E134A mutant HaNDK. The change from dimeric to tetrameric assembly is attributed to the removal of negative charge repulsion caused by the E134 in the wild-type HaNDK.