Order effect of base excision processes to repair clustered DNA damage
クラスターDNA損傷に対する塩基除去修復酵素の作用機序の効果
白石 伊世; 椎名 卓也; 菅谷 雄基  ; 鹿園 直哉; 横谷 明徳
; 鹿園 直哉; 横谷 明徳
Shiraishi, Iyo; Shiina, Takuya; Sugaya, Yuki; Shikazono, Naoya; Yokoya, Akinari
In a living cell, a multiply damaged site in DNA is thought to be processed by several different pathways simultaneously or sequentially. Under this situation the cellular response to the lesion cluster might depend on the order of repair processes because the configuration of the lesions will be modified by the reaction of initial repair protein, affecting the DNA-binding or lesion-excision activities of latter repair proteins. Plasmid DNA (pUC18) irradiated with C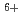 ion is treated with two base excision repair enzymes, Nth and Fpg, which convert pyrimidine and purine lesions to a SSB, respectively. Obtained results show that the amount of enzymatically induced SSB is very slightly less in DNA sample treated with Nth first and then Fpg than that of other treatments. These results indicate that the configurational change of the cluster by the first enzymatic treatment does not significantly influence the activity of secondary enzyme.
ion is treated with two base excision repair enzymes, Nth and Fpg, which convert pyrimidine and purine lesions to a SSB, respectively. Obtained results show that the amount of enzymatically induced SSB is very slightly less in DNA sample treated with Nth first and then Fpg than that of other treatments. These results indicate that the configurational change of the cluster by the first enzymatic treatment does not significantly influence the activity of secondary enzyme.