Crystal structure of endo-1,4- -glucanase from Eisenia foetida
-glucanase from Eisenia foetida
シマミミズ由来1,4- -エンドグルカナーゼの結晶構造解析
-エンドグルカナーゼの結晶構造解析
有森 貴夫*; 伊藤 彰紘*; 中澤 昌美*; 上田 光宏*; 玉田 太郎
Arimori, Takao*; Ito, Akihiro*; Nakazawa, Masami*; Ueda, Mitsuhiro*; Tamada, Taro
We cloned the gene for endo-1,4- -glucanase from Eisenia foetida (EF-EG2) consisting of 1368 bp encoding 456 amino acid residues. The amino acid sequence of the gene shares sequence homology (
-glucanase from Eisenia foetida (EF-EG2) consisting of 1368 bp encoding 456 amino acid residues. The amino acid sequence of the gene shares sequence homology (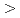 50%) with endo-1,4-
50%) with endo-1,4- -glucanases belonging to glycoside hydrolase family 9. The recombinant EF-EG2 showed highest activity at 40
-glucanases belonging to glycoside hydrolase family 9. The recombinant EF-EG2 showed highest activity at 40 C, and also retained a comparatively high activity at 10
C, and also retained a comparatively high activity at 10 C. We purified the recombinant EF-EG2 expressed in Pichia pastoris, and then grew needle shaped EF-EG2 crystals with dimensions of 0.02
C. We purified the recombinant EF-EG2 expressed in Pichia pastoris, and then grew needle shaped EF-EG2 crystals with dimensions of 0.02  0.02
0.02  1 mm. Diffraction data of EF-EG2 was collected at beamline 1A, Photon Factory, KEK, and integrated and scaled to 2.1
1 mm. Diffraction data of EF-EG2 was collected at beamline 1A, Photon Factory, KEK, and integrated and scaled to 2.1  resolution. The crystals belonged to space group
resolution. The crystals belonged to space group 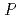 3
3 21 with unit-cell parameters of
21 with unit-cell parameters of 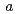 =
= 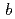 = 135
= 135  ,
, 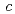 =54.9
=54.9  . The location of one EF-EG2 molecule in the asymmetric unit was identified by molecular replacement analysis using the coordinates of endoglucanase from Nasutitermes takasagoensis (NtEgl). The final model of EF-EG2 was refined to a crystallographic
. The location of one EF-EG2 molecule in the asymmetric unit was identified by molecular replacement analysis using the coordinates of endoglucanase from Nasutitermes takasagoensis (NtEgl). The final model of EF-EG2 was refined to a crystallographic 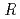 -factor of 17.9% (free R-factor of 21.4%) to 2.1
-factor of 17.9% (free R-factor of 21.4%) to 2.1  resolution. Overall structure of EF-EG2 is similar to that of NtEgl with an RMSD value of 0.9
resolution. Overall structure of EF-EG2 is similar to that of NtEgl with an RMSD value of 0.9  for 423 C
for 423 C atoms, however, a slight difference between both structures was confirmed around substrate binding site.
atoms, however, a slight difference between both structures was confirmed around substrate binding site.